Abstract
The complete mitochondrial genome of Cheliceroides longipalpis Zabka (Araneae: Salticidae) is a circular DNA molecule of 14,334 bp in length (GenBank accession number MH891570), and contains a standard set of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and an A + T-rich region. The gene orientation of C. longipalpis was similar to those found in other spider mitogenomes. The overall nucleotide composition is 36.5% A, 8.1% C, 12.9% G, and 42.5% T. Twenty reading frame overlaps and seven intergenic regions were found in the mitogenome of C. longipalpis. Thirteen tRNAs lack the potential to form the cloverleaf-shaped secondary structure. Ten PCGs start with ATN codons, two genes (cox2 and cox3) begin with TTG, and cox1 use AGA as initiation codon. Nine PCGs use usual termination codon of TAA or TAG, whereas the remaining four had an incomplete termination codon (T). The phylogenetic relationships of 14 spider mitogenomes based on the concatenated amino acid sequences of 13 PCGs showed that C. longipalpis is closely related to Plexippus paykulli, which is consistent with the traditional taxonomy.
Jumping spider, the common name for members of the family Salticidae, is one of the dominant spider groups and ∼5000 described species were recognized in the world (Maddison and Hedin Citation2003; Maddison et al. Citation2008). These spiders have a highly coordinated jumping ability, they could capture prey and transverse from plant to plant (Zabka Citation1985; Pan et al. Citation2014). In this study, the samples of jumping spider Cheliceroides longipalpis were collected from a broadleaf forest in Libo county (E107°57′, N25°17′), Guizhou Province, China. The specimen was deposited in the spider specimen room of Guiyang University with an accession number GYU-GZML-30.
The complete mitogenome of C. longipalpis (GenBank accession number MH891570) is a circular DNA molecule of 14,334 bp in length, and contains a standard set of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rrnL and rrnS), 21 transfer RNA genes (tRNAs), and an A + T region. The gene orientation of C. longipalpis was similar to those found in previously determined spider mitogenomes (Wang et al. Citation2016). Twenty-two genes were transcribed on the major strand (J-strand), whereas the others were oriented on the minor strand (N-strand). The nucleotide composition of C. longipalpis mitogenome is heavily biased toward A + T nucleotides, accounting for 36.5% A, 8.1% C, 12.9% G, and 42.5% T. The AT-skew and GC-skew of this genome were −0.075 and 0.232, respectively.
Gene overlaps in the C. longipalpis mitogenome were found in 20 locations and their total size is 282 bp. The longest overlap is 55 bp in length and located between trnS1 and trnR. This mitogenome has a total of 97 bp intergenic spacer sequences, which is made up of 7 regions in the range from 2 to 69 bp. The longest intergenic spacer is situated between trnN and trnA. The major A + T-rich region was located between trnQ and trnM genes with a length of 657 bp long, and the A + T content was 82.04%. Two rRNAs (rrnL and rrnS) were located between trnL1 and trnV, and between trnV and trnQ, respectively. The rrnL was 1037 bp in length with A + T content of 82.45%, and the rrnS was 697 bp in length with A + T content of 83.50%. The length of 22 tRNAs ranged from 48 bp (trnV) to 71 bp (trnY) and 9 of them were encoded on the minor strand. Thirteen tRNAs could not be folded into typical cloverleaf-shaped secondary structures. Among them, ten tRNA genes (trnW, trnK, trnD, trnL2, trnR, trnF, trnP, trnT, trnL1 and trnV) lost the TΨC-stem and three tRNAs (trnA, trnS1 and trnS2) lost the dihydrouracil (DHU) arm.
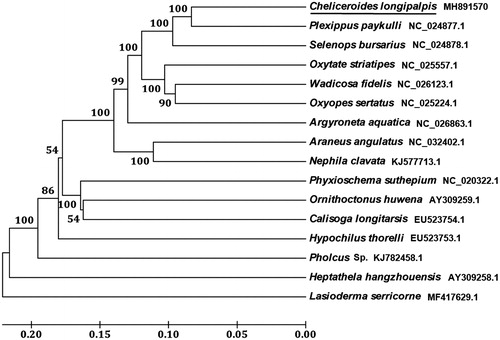
Ten PCGs in the C. longipalpis mitogenome start with ATN codons (ATG for nad3 and nad6; ATT for atp8; ATA for atp6, nad4, nad4L, nad5, cob, and nad2; ATC for nad1), two genes (cox2 and cox3) begin with TTG and cox1 use AGA as initiation codon. Nine PCGs use usual termination codon of TAA or TAG, whereas the remaining four PCGs had an incomplete termination codon (T). Based on the concatenated amino acid sequences of 13 PCGs, the neighbour-joining method was used to construct the phylogenetic relationship of C. longipalpis with 13 other representative spiders. The result showed that C. longipalpis is closely related to Plexippus paykulli (), which is consistent with the traditional taxonomy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Maddison WP, Hedin MC. 2003. Jumping spider phylogeny (Araneae: Salticidae). Invert Systematics. 17:529–549.
- Maddison WP, Bodner MR, Needham KM. 2008. Salticid spider phylogeny revisited, with the discovery of a large Australasian clade (Araneae: Salticidae). Zootaxa. 1893:49–64.
- Pan WJ, Fang HY, Zhang P, Pan HC. 2014. The complete mitochondrial genome of pantropical jumping spider Plexippus paykulli (Araneae: Salticidae). Mitochondrial DNA. 27:1–2.
- Wang ZL, Li C, Fang WY, Yu XP. 2016. The complete mitochondrial genome of two tetragnatha spiders (Araneae: Tetragnathidae): severe truncation of tRNAs and novel gene rearrangements in Araneae. Int J Biol Sci. 12:109–119.
- Zabka M. 1985. Systematic and zoogeographic study on the family Salticidae (Araneae) from viet-nam. Ann Zool Warszawa. 39:197–485.