Abstract
Taxillus nigrans (Hance) Danser (Loranthaceae) is a widely distributed mistletoe species in low mountains, hills, and river basins in subtropical areas of southwestern China. The whole chloroplast (cp) genome sequence of T. nigrans has been characterized using Illumina pair-end sequencing. The complete cp genome was 121,419 bp in length, containing a large single copy (LSC) region of 70,181 bp and a small single copy (SSC) region of 6100 bp, which were separated by a pair of 22,569 bp inverted repeat (IRs) regions. The genome contained 106 genes, including 66 protein-coding genes, 28 tRNA genes, 8 ribosomal RNA genes, and 4 pseudogenes. The most of gene species occur as a single copy, while 15 gene species occur in double copies. The overall GC content of T. nigrans cp genome is 37.4%, while the corresponding values of the LSC, SSC, and IR regions are 34.8, 26.3, and 42.95%, respectively. Phylogenetic analysis revealed that T. nigrans is closely related to T. sutchuenensis, with strong support values.
Taxillus nigrans (Hance) Danser (Loranthaceae) is a mistletoe species that is found attached to many canopy tree species in low mountains, hills, and river basins in subtropical areas of southwestern China at elevations of 300–1300 m. This species is widely used as raw material for Chinese traditional medicine (Jiang Citation1998). However, because the range of the species has undergone rapid expansion mediated by birds in the urban area of Chengdu (Sichuan Province, China), it forms large groves on garden tree species and is sometimes harmful to its host trees, so that individuals of this species are often removed by gardeners (Miao et al. Citation2017). A good knowledge of the genetic information of this species will help in researching the phylogeny of the Santalales order. In this study, we assembled and characterized the complete chloroplast (cp) genome sequence of T. nigrans based on the Illumina pair-end sequencing data.
The experimental samples were collected from the fresh leaves of T. nigrans on a host tree species, Platanus acerifolia in the Wangjiang campus of Sichuan University (E 104.08293°, N 30.63502°), and immediately stored in liquid nitrogen below −80 °C. Total genomic DNA was extracted with a modified CTAB method (Doyle Citation1987). A total of 22 million 150-bp raw pair-end reads were yielded by an Illumina Hiseq 2500 platform (Illumina, San Diego, CA). The Trimmomatic (Bolger et al. Citation2014) was used to filter the raw reads and get high-quality clean reads. We used NOVOPlasty v2.6.3 (Dierckxsens et al. Citation2017) and Velvet software (Zerbino and Birney Citation2008) to assemble and splice cp genomes using T. chinensis complete cp genome (Li et al. Citation2017) as a reference. Contigs were spliced into scaffold sequences and then used to assemble the cp genome. We used Geneious v11.0.3 (Kearse et al. Citation2012) to check the assembly results, and then we used Plann V1.1 (Huang and Cronk Citation2015) to refer to the results of T. chinensis cp genome annotation as a reference to annotate the T. nigrans cp genomes. The Geneious (Kearse et al. Citation2012) and Sequin V15.10 (http://www.ncbi.nlm.nih.gov/Sequin/) were used to correct the annotation results. The complete cp genome sequence was deposited in GenBank under accession number MH095982.
We used the online program Organellar Genome DRAW (OGDRAW) v1.2 (http://ogdraw.mpimp-golm.mpg.de/) to get the cp gene maps. To infer the phylogenetic relationships of Santalales, we constructed a phylogenetic tree based on another 10 Santalales species’ cp genomes. Cp genomes data for those 10 species were downloaded from the NCBI database, including Champereia manillana (NC_034931.1), Erythropalum scandens (NC_036759.1), Osyris alba (NC_027960.1), Schoepfia jasminodora (NC_034228.1), T. chinensis (NC_036306.1), T. sutchuenensis (NC_036307.1), Viscum album (NC_028012.1), V. coloratum (NC_035414.1), V. crassulae (NC_027959.1), and V. minimum (NC_027829.1).
The amino acid sequences of proteins encoded by their common genes were extracted and aligned using MAFFT V7.158 (Katoh and Standley Citation2013) and MEGA v6.0 (Tamura et al. Citation2013). We used the RAxML V8.2.11 software (Stamatakis Citation2014) to construct a ML tree. Two holoparasitic species of Ericales (Pylora rotundifolia KU833271.1 and V. macrocarpon NC_019616.1) were adopted as outgroups. Bootstrap analysis was performed with 1000 replicates. Then we used FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) to check the results.
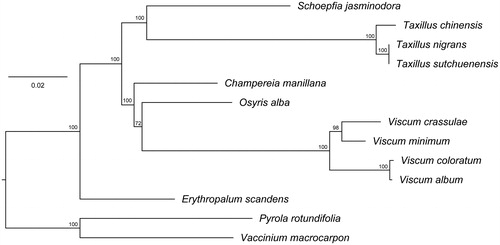
The T. nigrans cp genome is 121,419 bp in length, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 70,181 bp, two inverted repeats (IR) regions of 22,569 bp and a small single copy (SSC) region of 6100 bp. The genome contained 106 genes, including 66 protein-coding genes, 28 tRNA genes, 8 ribosomal RNA genes, and 4 pseudogenes. The most of gene species occur as a single copy, while 15 gene species occur in double copies. The overall GC content of T. nigrans cp genome is 37.4%, while the corresponding values of the LSC, SSC, and IR regions are 34.8, 26.3, and 42.95%, respectively. Phylogenetic analysis () demonstrated that 11 out of the 12 species of Santalales clustered into two highly-supported clades, Viscum (4 spp.) and Osyris (Santalaceae) plus Champereia (Opiliaceae) formed one clade, while Taxillus (3 spp.) and Scurrula (Loranthaceae) plus Schoepfia (Schoepfiaceae) formed another clade, whereas Erythropalum (Erythropalaceae) was the most distant clade within the Santalales. Although the monophyly of Viscum was strongly supported, the monophyly of Santalaceae, as implied by the apparent sister relationship between Viscum (Visceae) and Osyris (Santaleae), received a moderate bootstrap support (75%) and merits closer investigation.
Figure 1. Phylogenetic relationships of Santalales species using whole chloroplast genome. GenBank accession numbers: C. manillana (NC_034931.1), E. scandens (NC_036759.1), O. alba (NC_027960.1), S. jasminodora (NC_034228.1), T. chinensis (NC_036306.1), T. sutchuenensis (NC_036307.1), V. album (NC_028012.1), V. coloratum (NC_035414.1), V. crassulae (NC_027959.1), and V. minimum (NC_027829.1).
In summary, the complete cp genome from this study do not only provides important insight into its genome structure and composition but also plays a critical role in constructing phylogeny of the Santalales order.
Acknowledgements
The authors are grateful to the opened raw genome data from the public database.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. App Plant Sci. 3:1500026.
- Jiang ML. 1998. Flora Reipublicae Popularis Sinicae. Vol. 24. Beijing, China: Science Press. (in Chinese).
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li Y, Zhou J, Chen X, Cui Y, Xu Z, Li Y, Song J, Duan B, Yao H. 2017. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic taxillus species. Sci Rep. 7:12834.
- Miao N, Zhang L, Li M, Fan L, Mao K. 2017. Development of EST-SSR markers for Taxillus nigrans (Loranthaceae) in southwestern China using next-generation sequencing. App Plant Sci. 5:1700010.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
