Abstract
Non-monophyletic Poecilasmatids are deep-water epibiotic barnacles and are classified as five-capitular-plate members of the Lepadiformes. Here, we offer the first characterization of the mitogenome of a Poecilasmatid barnacle (Glyptelasma annandalei). The mitogenome was 16,107 bp long, with 72.3% AT content. Its gene arrangement was identical to that of the lepadids Lepas anserifera and L. australis, except for the location of the tRNASer gene. On the mitogenomic tree, two Lepadiformes families (Poecilasmatidae and Lepadidae) were monophyletic, which concurs with the findings of previous studies. Further mitogenomic analysis of undetermined Lepadiformes taxa is required to deepen our understanding of their phylogeny and evolution.
In traditional taxonomic classification, the stalked thoracican order Lepadiformes is composed of members having five capitular plates (Lepadomorpha) or reducing plates (Heteralepadomorpha; Buckeridge and Newman Citation2006). A previous molecular phylogenetic analysis found Lepadiformes to be monophyletic, with high bootstrap support (Pérez-Losada et al. Citation2008). However, most families belonging to Lepadiformes have been considered paraphyletic assemblages, with the exception of the Lepadidae. Additionally, the plesiomorphic shell plate state of the naked thoracican Heteralepadomorpha is considered a secondary loss in the evolution of stalked barnacles (Pérez-Losada et al. Citation2008).
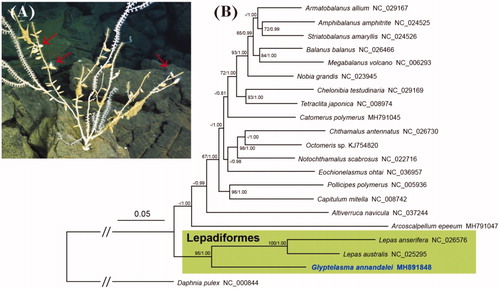
As of 11 September 2018, GenBank contains two complete mitochondrial genomes (mitogenomes) from lepadid barnacles of Lepadiformes, but no mitogenomes from the other families in this order. To understand the phylogenetic relationships and evolution of Lepadiformes, we determined the first mitogenome sequence of a deep-water epibiotic barnacle, Glyptelasma annandalei, in the family Poecilasmatidae. On 02 December 2016, G. annandalei was collected from the branches of a bamboo coral (Family Isididae) in the North Fiji Basin (17°07'S, 173°52'E; 2254 m depth). Genomic DNA extraction, sequencing, gene annotation, and phylogenetic analyses followed the methods of Kim et al. (Citation2017, Citation2018). The ambiguous regions between ND6 and ND2 were determined completely by Sanger sequencing and comparative mitogenomics. The specimen used for mitogenomic analysis has been deposited in the Biodiversity Research Museum, Academia Sinica, Taiwan (ASIZCR).
The complete mitogenome of G. annandalei was 16,107 bp in length (GenBank accession no. MH891848). It consisted of 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and a non-coding region. The gene organization followed the typical Pancrustacean gene arrangement and was identical to that of the lepadids Lepas anserifera (NC_026576) and L. australis (NC_025295), except for the location of tRNASer. In the Lepas mitogenome, tRNASer falls between the 12S rDNA and tRNALys genes, whereas it positions between CYTB and tRNAIle genes in G. annandalei.
The base composition was 38.0% A, 16.1% C, 11.5% G, and 34.3% T. All of the PCGs had ATN start codons (except ND2, which was initiated by a TTG codon) and TAA stop codons. The 16S and 12S rRNAs were 1307 bp (78.1% AT content) and 751 bp (73.2% AT content), respectively. The tRNA genes ranged from 56 to 70 bp in size. A 774-bp-long (79.8% AT content) non-coding region was located between the 12S rRNA and tRNALys.
Phylogenetic trees were constructed with the PCGs of 20 barnacles and a non-barnacle outgroup species, Daphnia pulex (NC_000844), using maximum likelihood (ML) and Bayesian inference (). These trees supported the monophyly of the poecilasmatid G. annandalei and two Lepas species, with high bootstrap support and Bayesian posterior probability; this concurs with previous studies based on nuclear and partial mitochondrial genes (Pérez-Losada et al. Citation2008; Herrera et al. Citation2015). However, taxonomic ambiguities among poecilasmatid, lepadid, and heteralepadid members still remain. The mitogenomes of the Lepadiformes families Heteralepadidae, Koleolepadidae, and Oxynaspididae have not been determined, and further mitogenomic analysis of undetermined taxa is required to deepen our understanding of their phylogeny and evolution.
Figure 1. (A) Photograph of the sampling site in the North Fiji Basin where the epibiotic barnacle Glyptelasma annandalei was collected from the branches of a bamboo coral (Family Isididae). Arrows indicate G. annandalei individuals. (B) Phylogenetic tree of G. annandalei and other thoracican barnacles based on 13 mitochondrial protein-coding genes. The model GTR + I + G was selected as the best evolutionary model using jModelTest 2.1.4. The green-shaded box contains two families of the order Lepadiformes. Numbers at internodes are the maximum likelihood bootstrap proportions (left) and Bayesian posterior probabilities (right). An asterisk indicates a bootstrap value of less than 60%.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and composition of this paper.
Additional information
Funding
References
- Buckeridge JS, Newman WA. 2006. A revision of the Iblidae and the stalked barnacles (Crustacea: Cirripedia: Thoracica), including new ordinal, familial and generic taxa, and two new species from New Zealand and Tasmanian waters. Zootaxa. 1136:1–38.
- Herrera S, Watanabe H, Shank TM. 2015. Evolutionary and biogeographical patterns of barnacles from deep-sea hydrothermal vents. Mol Ecol. 24:673–689.
- Kim S-J, Lee W-K, Hou BK, Chan BKK, Ju SJ. 2017. Complete mitochondrial genome of the deep-sea asymmetrical barnacle Altiverruca navicula (Cirripedia, Thoracica, Verrucumorpha). Mitochondrial DNA B Resour. 2:934–935.
- Kim S-J, Lee W-K, Kim RO, Ju SJ. 2018. Complete mitochondrial genome of the hydrothermal vent barnacle Eochionelasmus ohtai (Cirripedia, Thoracica). Mitochondrial DNA B Resour. 3:46–47.
- Pérez-Losada M, Harp M, Høeg JT, Achituv Y, Jones D, Watanabe H, Crandall KA. 2008. The tempo and mode of barnacle evolution. Mol Phylogenet Evol. 46:328–346.
