Abstract:
Leontice incerta, which belongs to Berberidaceae, is an endangered species in China. In this study, the chloroplast (cp) genome of L. incerta was assembled using genome skimming sequencing and the phylogeny of Berberidaceae was reconstructed based on whole cp genome. The cp genome of L. incerta is 156,923 bp in length, comprising two copies of IR (26,121 bp) regions separated by the LSC (85,622 bp) and SSC (19,059 bp) regions. The cp genome encodes 112 unique genes, consisting of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Phylogenetic analysis indicates that L. incerta is sister to Gymnospermium microrrhynchum, subsequently is sister to Nandia domestica.
Leontice L. is a small perennial, tuberous and herbaceous genus in Berberidaceae, containing merely four species named L. armeniaca Belanger, L. ewersmanni Bunge, L. incerta Pall. and L. leontopetalum L. (Wu et al., Citation2011). Among these species, L. incerta is the only species distributed in China and is native to Xinjiang. L. incerta is now considered as an endangered species and listed on the list of key protected wild plants by Xinjiang Provincial Government (Citation2007). However, the studies regarding to the genetic background of L. incerta are scarce up to now, except some karyological studies (Kosenko Citation1977). In this study, the complete chloroplast genome of L. incerta was determined using genome skimming data. The complete chloroplast genome sequence was registered into GenBank with the accession number MH940295.
Genomic DNA was extracted from silica-dried leaf tissue of one L. incerta plant collected in Xinjiang (China; 85°52′17.04″E, 44°18′24.12″N) using Plant DNAzol Reagent (LifeFeng, Shanghai) according to the manufacturer’s protocol. A voucher specimen (Pan Li LP173506) was deposited at the Herbarium of Zhejiang University (HZU). The high quality DNA was sheared and the libraries preparation and sequencing were performed on Illumina HiSeq 2500 by Beijing Genomics Institute (Wuhan, China). The raw data were filtered by quality with Phred score <30 and the remaining sequences were assembled into contigs using the CLC De novo assembler beta 4.06 (CLC Inc., Rarhus, Denmark) to reconstruct the cp genome with Gymnospermium microrrhynchum (GenBank accession number: NC_030061) as a reference. The annotation of the cp genome was performed through the online program Dual Organellar Genome Annotator (Wyman et al. Citation2004), and the circular gene maps were drawn by the OrganellarGenomeDRAW tool (OGDRAW) following by manual modification (Lohse et al. Citation2013). The phylogenetic tree was reconstructed based on whole cp genome sequences of 27 Berberidaceae species using maximum likelihood (ML) method, with two Ranunculus species as outgroup. ML analysis was implemented in RAxML-HPC v8.1.11 on the CIPRES cluster1 (Miller et al. Citation2010).
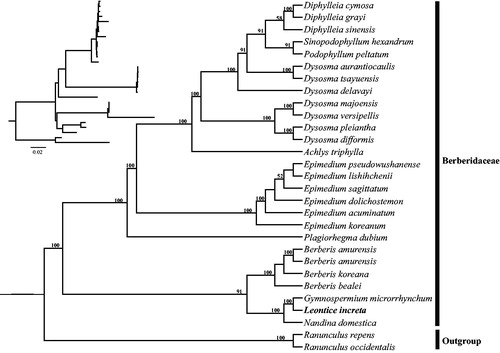
The complete chloroplast genome of L. incerta is 156,923 bp in length and shared the common feature of comprising two copies of IR (Inverted region, 26,121 bp) regions separated by the LSC (Large single copy, 85,622 bp) and SSC (Small single copy, 19,059 bp) regions. The overall GC content of the total length, LSC, SSC, and IR regions is 38.3%, 36.5%, 32.5%, and 43.2%, respectively. Within the chloroplast genome of L. incerta there are 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, four rRNA genes, with 19 duplicated genes in the IR regions. Six tRNA genes and nine protein-coding genes contain single intron, and three genes including rps12, clpP and ycf3 contain two introns. The phylogeny revealed that L. incerta and G. microrrhynchum formed a clade with high support, which is sister to Nandina domestica ().
Figure 1. Phylogenetic tree reconstruction of Berberidaceae using maximum likelihood (ML) based on whole chloroplast genome sequences. Relative branch lengths are indicated at the top-left corner. Numbers above the lines represent ML bootstrap values. ML bootstrap <50% was not shown. GenBank accession numbers of taxa are shown below, Nandina domestica (DQ923117), Berberis amurensis (KM057377), Berberis bealei (NC_022457), Berberis amurensis (NC_030062), Berberis koreana (NC_030063), Sinopodophyllum hexandrum (KT445939), Epimedium sagittatum (NC_029428), Epimedium acuminatum (NC_029941), Epimedium dolichostemon (NC_029942), Epimedium koreanum (NC_029943). Epimedium lishihchenii (NC_029944), Epimedium pseudowushanense (NC_029945), Gymnospermium microrrhynchum (NC_030061), Ranunculus occidentalis (NC_031651), Ranunculus repens (NC_036976), Achlys triphylla (NC_037726), Dysosma versipellis (NC_037898), Dysosma delavayi (NC_037899), Dysosma majoensis (NC_037900), Dysosma aurantiocaulis (NC_037902), Dysosma tsayuensis (NC_037904), Dysosma pleiantha (NC_037905), Dysosma difformis (NC_037906), Diphylleia grayi (NC_037901), Diphylleia sinensis (NC_037907), Diphylleia cymosa (NC_037908), Plagiorhegma dubium (NC_038103).
In conclusion, the complete chloroplast genome of L. incerta is reported for the first time in this study. It will provide essential and important genetic resources for future conservation of this endangered species, and also gain insights into evolutionary patterns within the Berberidaceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kosenko VN. 1977. Comparative karyological study of Leontice ewersmanni Bunge and Leontice incerta Pall. (Berberidaceae). Bot Z.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575.
- Miller M-A, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). 2010. IEEE. 1–8.
- Wu C, Raven P, Hong D. 2011. Flora of China. Vol. 19 (Cucurbitaceae through Valerianaceae, with Annonaceae and Berberidaceae). Science Press/Missouri Botanical Garden Press, Beijing, China/St. Louis, MO, USA.
- Wyman S-K, Jansen R-K, Boore J-L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xinjiang Provincial Government. 2007. Provincial Key Protected Plant List. (accessed 2007 August 27). http://www.chinalawedu.com/falvfagui/fg22016/251457.shtml.
