Abstract
The mitochondrial genome of the Encyrtus infelix was sequenced and it is the first completed mitochondrial genome of species in the family Encyrtidae (Hymenoptera: Chalcidoidea). This mitochondrial genome is 15,698 bp in length with 37 classical eukaryotic mitochondrial genes and an A + T-rich region. All the 13 PCGs begin with typical ATN codons. Among them, seven genes terminate with TAA, one with TA– and five with T––. All of the 22 tRNA genes range from 56 to 70 bp with typical cloverleaf structure except for trnS1, whose dihydrouridine (DHU) arm forms a simple loop. Gene directions and arrangements are similar to Eurytoma sp. mitochondrial genome. Encyrtus infelix also has a dramatic gene rearrangement, the placements of some genes are peculiar to Chalcidoidea. Phylogenetic analysis highly supported the monophyly of both Chalcidoidea and Pteromalidae.
Encyrtidae is one of the largest families of superfamily Chaidoidea (Insecta: Hymenoptera), with currently over 4000 species in 490 genera (Noyes Citation2018). They attack a wide range of insect hosts, other from ticks and spiders. They are extremely important as biological control agents. However, no complete or nearly complete mitogenome of any species in this family has been reported. Here, we present the first complete mitogenome of Encyrtus infelix (Embleton 1902), a species of the type genus of family Encyrtidae. Specimens of E. infelix were reared from Saissetia hemisphaerica (Hemeptera: Coccidae) collected in Kunming, Yunnan. Now the specimens are stored in the Institute of Zoology, Chinese Academy of Sciences (voucher number: E3-162A, E3-326A).
The complete sequence of E. infelix was 15,698 bp in length with A + T content of 78.4%, containing 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNA), 2 ribosomal RNAs (rRNA), and a putative control region (CR). Analysis of the E. infelix complete mitochondrial genome revealed dramatic mitochondrial gene rearrangement, in which gene directions and rearrangements are similar to Eurytoma sp. mitochondrial genome (Su et al. Citation2016). Additionally, 17 intergenic spacers (351 bp in total) and 7 overlapping regions (20 bp in total) are dispersed throughout the genome.
All 13 PCGs were initiated by typical ATN codons (seven ATT, five ATG, and one ATA). Seven genes terminate with TAA, six genes have incomplete stop codon. All of the 22 tRNA genes, ranging from 56 to 70 bp, have a typical cloverleaf structure except for trnS1, whose dihydrouridine (DHU) arm forms a simple loop. A lack of the DHU arm in trnS1 was found in the mitochondrial genomes of most insects (Wolstenholme Citation1992) and other metazoans (Lavrov et al. Citation2000). The control region was 709bp long and 84.5% A + T content. The rrnL and rrnS genes are 1291 and 757 bp, with an average A + T content of 84.7% and 82.6%, respectively. Both of them are located on the minority stand and separated by trnA and trnQ, which is unique in the available hymenopterans mitochondrial genomes.
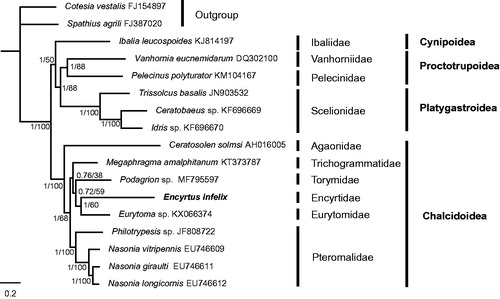
We selected 14 complete or nearly complete mitogenomes of Proctotrupomorpha whose mitochondrial genome were available in NCBI. Two species from superfamily Ichneumonoidea, Cotesia vestalis and Spathius agrili were used as outgroup. We conducted phylogenetic analyses based on all PCGs, even there were incomplete PCGs in some species using both MrBayes (Ronquist et al. Citation2012) and raxmlGUI (Silvestro and Michalak Citation2012). Two consensus trees displayed the identical topology with high supports for most nodes (). According to the phylogenetic tree, the monophyly of Chalcidoidea was strongly supported. Within Chalcidoidea, our analysis supported the basal position of Agaonidae and its monophyly, Pteromalidae was strongly supported as monophyletic, which is in agreement with previous analyses (Yang et al. Citation2018). Additional sampling taxa of Chalcidoidea are needed to obtain robust phylogenetic tree for the better understanding of the evolutionary history of Chalcidoidea.
Figure 1. The phylogenetic tree of the tested Proctotrupomorpha and two outgroups using 13 PCGs sequences of mitochondrial genomes. The numbers at the nodes separated by “/” indicates the Bayesian posterior probabilities (former) and raxmlGUI posterior probabilities (latter), respectively. Each species involved in the tree has scientific name on the right side.
Nucleotide sequence accession number
The two mitogenomes sequences of Encyrtus infelix have been assigned. GenBank accession numbers are MH574908 and MH729198.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the paper, and report no conflicts of interest.
Additional information
Funding
References
- Embleton AL. 1902. On the economic importance of the parasites of Coccidae. Trans Royal Entomol Soc London. 50:219–229.
- Lavrov DV, Brown WM, Boore JL. 2000. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA. 97:13738–13742.
- Noyes JS. 2018. Universal Chalcidoidea database. World Wide Web electronic publication. [accessed 2018 June 1]. http://www.nhm.ac.uk/chalcidoids.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Su TJ, Huang DY, Wu YP, He B, Liang AP. 2016. Sequencing and characterization of mitochondrial genome of Eurytoma sp. (Hymenoptera: Eurytomidae). Mitochondrial DNA Part B. 1:826–828.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173.
- Yang J, Liu HX, Li YX, Wei ZM. 2018. The rearranged mitochondrial genome of Podagrion sp. (Hymenoptera: Torymidae), a parasitoid wasp of mantis. Genomics. https://doi.org/10.1016/j.ygeno.2018.02.020
