Abstract
Eurycoma longifolia is a small tree known for its use in Southeast Asian traditional medicine. The growing demand for its roots as raw material for medicinal extracts is slowly threatening the natural populations of this species. In this study, we assembled and characterized the complete chloroplast genome of E. longifolia as a resource for future genetic studies on this species. With a total length of 160,800 bp, comprising of a large single-copy (LSC) region of 88,162 bp, a small single-copy (SSC) region of 15,607 bp, separated by two inverted repeat (IR) regions of 27,369 bp each, 133 genes were predicted for the chloroplast genome. Phylogenetic analysis confirmed the position of E. longifolia within the family Simaroubaceae.
Eurycoma longifolia Jack is a small tree species that belongs to a three-member genus within the family Simaroubaceae. It is distributed in Southeast Asia, mainly in Malaysia, Indonesia and Vietnam, with small patches in other countries (Rehman et al. Citation2016). Although traditionally used as folk medicine in treatments of malaria, diabetes, cancer, etc. (Bhat and Karim Citation2010; Rehman et al. Citation2016), the recent popularity of the species has been attributed to aphrodisiac properties found in its root extract (Rehman et al. Citation2016). Nowadays, root extracts of E. longifolia are widely available as additives in everyday consumer products, especially in coffee and tea preparations, as well as health supplements. Due to popular demand, natural populations of E. longifolia face increasing harvesting pressure. In this study, we characterized the complete chloroplast genome sequence of E. longifolia as a resource for future genetic studies on this and other related species.
Fresh leaves were obtained from an E. longifolia plant purchased from a nursery in Kuala Lumpur, Malaysia. A voucher specimen (NG99001) is deposited at the herbarium of the China-ASEAN College of Marine Sciences, Xiamen University Malaysia. After DNA extraction, a library with insertion size of ∼400 bp was constructed, and high-throughput DNA sequencing (pair-end 150 bp) was performed on an Illumina HiSeq 2500 platform. Approximately 4 Gb of sequence data was generated. Using a E. longifolia partial atpB gene sequence (EU042787.1) as seed and the Leitneria floridana chloroplast genome sequence (KT692940.1) as a reference, the chloroplast genome was assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome sequence was then annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of E. longifolia (GenBank accession MH751519) was 160,800 bp in length, with a large single-copy (LSC) region of 88,162 bp and a small single-copy (SSC) region of 15,607 bp, separated by a pair of inverted repeat (IR) regions of 27,369 bp each. A total of 133 genes were predicted, consisting of 89 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content was 37.64%.
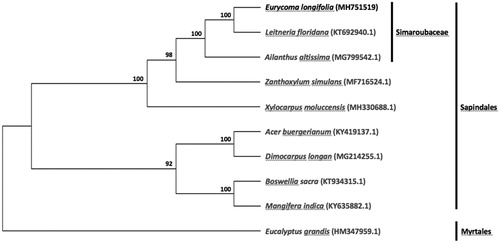
To understand the relationship of E. longifolia within the family Simaroubaceae and within the order Sapindales, chloroplast genome sequences of eight other representative species from the order Sapindales (including two from the family Simaroubaceae) and Eucalyptus grandis from the order Myrtales were downloaded from the NCBI GenBank database for phylogenetic analysis. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and then, RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with E. grandis as outgroup. As shown in , E. longifolia was found to be clustered within Simaroubaceae with strong bootstrap support.
Figure 1. Maximum likelihood tree showing the relationship among Eurycoma longifolia and representative species within the order Sapindales, based on whole chloroplast genome sequences, with Eucalyptus grandis of the order Myrtales as outgroup. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Bhat R, Karim AA. 2010. Tongkat Ali (Eurycoma longifolia Jack): a review on its ethnobotany and pharmacological importance. Fitoterapia. 81:669–679.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Rehman SU, Choe K, Yoo HH. 2016. Review on a traditional herbal medicine, Eurycoma longifolia Jack (Tongkat Ali): its traditional uses, chemistry, evidence-based pharmacology and toxicology. Molecules. 21:331.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
