Abstract
Ulmus chenmoui is of great ecological and horticultural importance and a national Class III protected plant endemic to China. However, previous studies of it suffered from insufficient genomic resources. Here, using the paired-end shotgun sequencing technology, we determined the complete chloroplast (cp) genome sequence of U. chenmoui. Our results showed the cp genome of U. chenmoui was 159,371 bp long and displayed a typical quadripartite structure which consisted of a pair of inverted repeats (IRs) with a length of 26,296 bp, separating by two single-copy regions (LSC, 88,084 bp and SSC, 18,695 bp). Besides, the cp genome of U. chenmoui contained 120 genes, including 83 protein coding genes, 29 transfer RNAs and eight ribosomal RNAs. Moreover, a maximum likelihood (ML) phylogenetic analysis based on the 13 cp genomes demonstrated the monophyly of Ulmaceae and U. chenmoui was closely related to Ulmus gaussenii.
Ulmus chenmoui W. C. Cheng, which belongs to the eudicot family Ulmaceae sensu APG IV (The Angiosperm Phylogeny Group, Citation2016), is a drought-resistant deciduous tree growing in the limestone areas (Fu and Liu, Citation2001; Han et al. Citation2010). It narrowly distributes in Langya Mountain (Anhui, China) and Baohua Mountain (Jiangsu, China) (Hao et al. Citation2000). However, due to the over-exploitation for forestation and habitat destruction, the population size of U. chenmoui has sharply decreased, and thus in urgent need of protection (Geng et al. Citation2016; Liu et al. Citation2017). Additionally, U. chenmoui has been categorized as a National Key Protected Species in China (Class III) and classified as ‘Endangered’ by the IUCN (International Union for Conservation of Nature, http://www.iucnredlist.org/). In recent years, with the advent of next-generation sequencing technologies, abundant genomic resources of our studied species have been produced in a rapid and cost-effective way ( Yu et al. Citation2011; Malé et al. Citation2014). In this study, we characterized the complete cp genome sequence of U. chenmoui based on the Illumina short-gun sequencing data and registered it into the GenBank with the accession number MH704468. These newly available genomic resources here will greatly promote the further conservation genetic studies of this endangered species.
Total genomic DNA was extracted from silica-dried leaves of one wild U. chenmoui plant sampled from Langya Mountain (China; 32°16.5506'N, 118°16.5685'E) using a modified CTAB method (Yang et al. Citation2014) and the voucher specimen was deposited at the Herbarium of Zhejiang University (HZU). Paired-end (2 × 125 bp) sequencing library was constructed and sequenced on the Illumina HiSeq2500 platform at Beijing Genomics Institute (Shenzhen, China). Approximately 2.5 Gb of raw data were firstly trimmed to remove low-quality reads (Q < 20, .01 probability error) and adapter sequences using the CLC-quality trim tool. Filtered reads were then assembled into contigs using CLC Genomics Workbench v8.5.1. All the high-quality contigs were aligned to the reference cp genome of Ulmus davidiana (KY244082; Zuo et al. Citation2017) via BLAST (http://blast.ncbi.nlm.nih.gov/) and the draft cp genome of U. chenmoui was constructed by connecting the overlapping terminal sequences in Geneious v11.0.4 (http://www.geneious.com/). Subsequently, clean reads were remapped to the draft cp genome and yielded the final cp genome sequence of U. chenmoui. Gene annotation was performed using the Dual Organellar GenoMe Annotator (Wyman et al. Citation2004), and the circular cp physical map representing U. chenmoui was drawn by OrganellarGenome DRAW (Lohse et al. Citation2007) with manual editing.
The complete cp genome of U. chenmoui was 159,371 bp long and it displayed the typical quadripartite structure of nearly all land plants, consisting of a pair of inverted repeat regions (IRs with 26,296 bp) separated by two single-copy regions (LSC, 88,084 bp and SSC, 18,695 bp). The overall GC content of the cp genome was 35.6%, with the corresponding values of 33.0, 32.6 and 42.4% for the LSC, SSC and IR regions, respectively. Besides, the cp genome of U. chenmoui encoded 120 predicted functional genes, of which 15 were duplicated in the IR regions. Among the unique genes, there were 75 protein-coding genes (CDS), 24 tRNA genes and four rRNA genes. Five protein-coding genes and one tRNA gene contained a single intron, while three protein-coding genes possessed two introns. The gene rps12 was trans-spliced; with the 5′-end exon located in the LSC region and two copies of 3′-end exon and intron in the IR regions. Moreover, the ψatpF, ψclpP and ψycf15 were identified as pseudogenes because of the partial duplication.
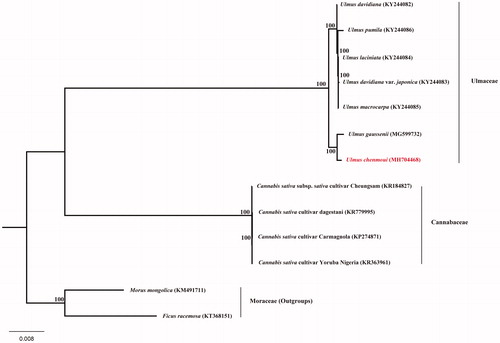
Furthermore, we reconstructed the phylogeny of Ulmaceae based on the previously reported complete cp genomes of related taxa, employing the GTR + R + I model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference in RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. Citation2010). Our phylogenetic tree showed the seven species of Ulmaceae formed a monophyletic clade with full support at all the nodes and U. chenmoui was closely related to Ulmus gaussenii (). These genomic resources and plastid phylogenomics here will largely enrich the genetic resources and enhance the conservation genetics of U. chenmoui.
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Fu SL, Liu SQ. 2001. Drought-relief characters of several species on limestone area. J Soil Water Conserv. 19:11–15.
- Geng QF, He J, Yang J, Shi E, Wang DB, Xu WX, Jeelani N, Wang ZS, Liu H. 2016. Development and characterization of microsatellite markers for Ulmus chenmoui (Ulmaceae), an endangered tree endemic to eastern China. Genet Mol Res GMR. 15:1–7.
- Han X, Zhi YB, Zhou ZZ, Sun ZY, Yang SB, Chen QN, Li JM. 2010. Study on pollen morphology and its geohistoric significance of two endangered elms peculiar to Mt. Langya, China. Acta Micropalaeontol Sin. 27:77–84.
- Hao RM, Huang ZY, Liu XJ, Wang ZL, Xu HQ, Yao ZG. 2000. The natural distribution and characteristics of the rare and endangered plants in Jiangsu, China. Chin Biodivers. 8:153–162.
- Liu L, Chen W, Yan DT, Li J, Liu L, Wang YL. 2017. Molecular phylogeography and paleodistribution modeling of the boreal tree species Ulmus lamellosa (T. Wang et SL Chang) (Ulmaceae) in China. Tree Genet Genomes. 13:11.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Malé PG, Bardon L, Besnard G, Coissac E, Delsuc F, Engel J, Lhuillier E, Scottisaintagne C, Tinaut A, Chave J. 2014. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Mol Ecol Res. 14:966–975.
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE). New Orleans, LA: IEEE; p.1–8.
- The Angiosperm Phylogeny Group, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 14:1024–1031. doi:10.1111/1755-0998.12251
- Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, et al. 2011. Next-generation sequencing to generate interactome datasets. Nat Methods. 8:478–480.
- Zuo LH, Shang AQ, Zhang S, Yu XY, Ren YC, Yang MS, Wang JM. 2017. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: genome comparative and taxonomic position analysis. PloS One. 12:e0171264.