Abstract
The complete chloroplast genome from Biondia insignis Tsiang, a rare liana of the Apocynaceae endemic to China is determined in this study. The whole chloroplast genome sequence of Biondia insignis Tsiang has been characterized by Illumina pair-end sequencing. The circular genome is 160,381 bp long, containing a large single copy region (LSC) of 91,493 bp and a small single copy region (SSC) of 19,262 bp, which are separated by a pair of 24,813 bp inverted repeat regions (IRs). It encodes a total of 127 genes, including 73 protein-coding genes (70 PCG species), 46 tRNA genes (31 tRNA species), and eight ribosomal RNA genes (4 rRNA species). The most of gene species occur as a single copy, while 14 gene species occur in double copies. The overall A + T content is 62.2%, while the corresponding values of the LSC, SSC, and IR regions are 63.9, 68.0, and 56.7%, respectively. The evolutionary relationships revealed by phylogenetic analysis indicated that Biondia insignis Tsiang was relatively close to Vincetoxicum rossicum. These complete chloroplast genomes can be subsequently used for population genomic studies of Biondia and provide valuable insight into conservation and exploitation for this rare species.
Biondia insignis Tsiang is an endemic liana of Apocynaceae mainly distributed in south-western China and occur in mountain forests at altitudes of 2000–2900 m (Agendae Academiae Sinicae Edita Citation1977). This plant and its related species contain most characteristic metabolites C21-steroidal compounds which have many potential medicinal values and biological activity (Shen et al. Citation2000; Zhang Citation2000). However, the precise phylogenetic position for this rare species of Biondia genera still remains uncertain. Analysing the chloroplast/plastid genome proved to be an efficient approach to shed light on plant molecular systematics (Moore et al. Citation2010; Zhang et al. Citation2011). In this study, we assembled and characterized the complete chloroplast genome sequence of Biondia insignis Tsiang based on the Illumina pair-end sequencing data, which is an important resource for future studies on development of molecular markers to monitor population dynamics of the rare species and further contribute to the formulation of conservation and exploitation strategy.
Genomic DNA was extracted from fresh leaves of an individual of Biondia insignis Tsiang collected from the rock bushes along the road of Heishui counties (102°56 49, 32°6; N; the specimen was deposited at Biotechnology Research Institute, CAAS, Beijing, China; accession number: GM-2018-44). The genomic library was sequenced on an Illumina Hiseq X Ten platform. The high-quality reads were assembled into complete chloroplast genomes using Velvet (Zerbino and Birney Citation2008) and filled gap with GapCloser (http://soap.genomics.org.cn/index.html). The assembled chloroplast genome was annotated using the online annotation tool DOGMA (Wyman et al. Citation2004) and further corrected the annotation with Geneious (Kearse et al. Citation2012), then submitted to GenBank (Accession No. MH748558).
The chloroplast sequence of the Biondia insignis Tsiang was 160,381 bp. The large single copy region (LSC) in Biondia insignis Tsiang was 91,493 bp and the small single copy region (SSC) was 19,262 bp, which were separated by a pair of 24,813 bp inverted repeat regions (IRs). The circular genome contained 127 genes, including 73 protein-coding genes (70 PCG species), eight ribosomal RNA genes (4 rRNA species) and 46 tRNA genes (31 tRNA species). The most of gene species occurred in a single copy, while 14 gene species occurred in double copies, including four rRNA species (4.5S, 5S, 16S, and 23S rRNA), 7 tRNA species, and 5 PCG species. The overall A + T content of the circular genome was 62.2%, while the corresponding values of the LSC, SSC, and IR regions were 63.9, 68.0, and 56.7%, respectively.
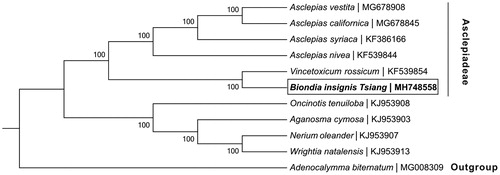
A neighbour-joining phylogenetic tree was constructed based on the chloroplast genomes for a panel of 11 species () with the program MEGA6 version 6 (Tamura et al. Citation2013). The phylogenetic analysis supported the traditional taxonomy of the family Apocynaceae. Furthermore, Biondia insignis Tsiang was found to be relatively closely related to Vincetoxicum rossicum and these two species were then clustered into a clade with other four Asclepiadeae species, while the rest of the Apocynaceae family clustered into another clade (). We believe this determined B. insignis chloroplast genome will provide useful resources for population genomic studies of Biondia and/or Asclepiadeae.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Agendae Academiae Sinicae Edita. 1977. Flora reipublicae popularis sinicae. Vol. 63. Tomus: Science Press Beijing; p. 397.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107:4623–4628.
- Shen YM, Zhang YH, Wen YY, Kuang TY. 2000. Insignin A, a novel C21-steroidal aglycone from Biondia insignis. Chin Chem Lett. 11:1065–1068.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang YH. 2000. The study of the C21 steroidal constituents of three species from Asclepiadaceae. [doctoral dissertation]. Beijing: Institute of Botany, the Chinese Academy of Sciences.
- Zhang YJ, Ma PF, Li DZ. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One. 6:e20596.