Abstract
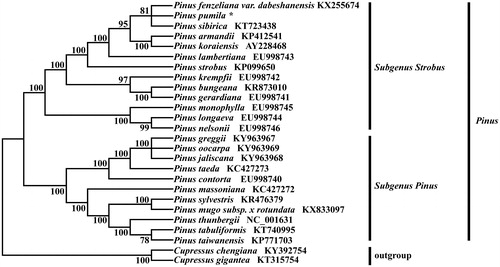
Pinus pumila is a rare and endangered conifer plant in the family Pinaceae. Here, we determined the complete chloroplast (cp) genome sequence of P. pumila. The cpDNA was 117,398 bp in length, containing a pair of 475 bp inverted repeat regions (IR), which was divided into large single copy region (LSC) and small single copy region (SSC) with 64,606 and 51,842 bp in length, respectively. The GC content in cpDNA, LSC region, SSC region and IR region were 38.8%, 38.0%, 39.8% and 37.9%, respectively. The cpDNA contained 114 genes, including 74 protein-coding genes (CDS), 36 transfer RNA genes (tRNA) and 4 ribosomal RNA genes (rRNA). A phylogenetic analysis revealed that P. pumila is more closely related with congeneric P. fenzeliana var. dabeshanensis and P. sibirica.
Pinus pumila is a rare and endangered plant in the family Pinaceae, which distributed in Northeast China, Japan and Eastern Siberia. Previous studies have focused on the genetic structure, paleoclimatic implications, vegetation dynamics, photosynthesis and respiration for this species (Okitsu and Ito Citation1984; Kajimoto Citation1990; Watano et al. Citation2004; Anderson et al. Citation2010; Chen et al. Citation2016). In this study, we sequenced and assembled the complete chloroplast genome (cpDNA) of P. pumila. The annotated cpDNA of P. pumila has been deposited into the GenBank with the accession number number MH046871.
Fresh leaf tissues were collected in the Greater Hinggan mountains, Heilongjiang Province, China (N: 51.62°; E: 124.00°; H: 627m). The total genomic DNA was extracted using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). DNA sample and voucher specimen (No. PPLZH2016137) of P. pumila were deposited in the Northwest University Museum (NUM). Meanwhile, we constructed a pair-end (PE) library with 350 bp insert size fragments using TruSeq DNA sample preparation kits (Sangon, Shanghai, China). Thereafter, we sequenced at least 1.2 GB of clean data for P. pumila. The detailed next-generation sequencing was conducted on the Illumina Hiseq 2500 platform by Sangon Biotech (Shanghai, China). Then, we trimmed the low-quality reads by NGSQCToolkit v2.3.3 (Patel and Jain Citation2012). After removing the low-quality sequences, the clean reads were assembled using MIRA v4.0.2 (Chevreux et al. Citation2004) and MITObim v1.8 (Hahn et al. Citation2013) with the cp genome of Pinus armandii (KP412541), as reference. A total of 208,396 bp reads generated with average coverage of 266.7. Then, we used DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004) and Geneious v8.0.2 (Kearse et al. Citation2012) to annotate the chloroplast genome. Finally, we acquired the P. pumila complete chloroplast genome sequences.
The complete chloroplast (cp) genome sequence of P. pumila was 117,398 bp in length, containing a pair of 475 bp inverted repeat regions (IR), one large single copy region (LSC) with 64,606 bp in length and one small single copy region (SSC) of 51,842 bp in length. The GC content in cpDNA, LSC region, SSC region and IR region were 38.8%, 38.0%, 39.8% and 37.9%, respectively. The cpDNA contained 114 genes, including 74 protein-coding genes (CDS), 36 transfer RNA genes (tRNA) and 4 ribosomal RNA genes (rRNA).
Twenty-five available complete chloroplast genomes, 23 Pinus species with Cupressus chengiana (KY392754) and Cupressus gigantean (KT315754) as outgroups, all of the 26 complete chloroplast sequences were aligned using MAFFT (Katoh and Standley Citation2013) with the default parameters. We constructed a maximum likelihood phylogenetic tree based on these complete plastomes using MAGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates under the GTRGAMMA model, which was determined using the Modeltest v3.7 (Posada and Crandall Citation1998). The phylogenetic analysis showed that the most of monophyletic clade with high bootstrap value (). P. pumila is more closely related with P. fenzeliana var. dabeshanensis and P. sibirica. Meanwhile, the P. pumila chloroplast genome will provide information for the conservation, utilization and management.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Anderson PM, Lozhkin AV, Solomatkina TB, Brown TA. 2010. Paleoclimatic implications of glacial and postglacial refugia for Pinus pumila in western Beringia. Quater Res. 73:269–276.
- Chen F, Zhang Q, Gu H, Yang L. 2016. An approach for extraction of kernel oil from Pinus pumila using homogenate-circulating ultrasound in combination with an aqueous enzymatic process and evaluation of its antioxidant activity. J Chromatogr A. 1471:68–79.
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kajimoto T. 1990. Photosynthesis and respiration of Pinus pumila needles in relation to needle age and season. Ecol Res. 5:333–340.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Boil Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Okitsu S, Ito K. 1984. Vegetation dynamics of the siberian dwarf pine (Pinus pumila, regel) in the taisetsu mountain range, hokkaido, japan. Vegetatio. 58:105–113.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Watano Y, Kanai A, Tani N. 2004. Genetic structure of hybrid zones between Pinus pumila and P. parviflora var. pentaphylla (pinaceae) revealed by molecular hybrid index analysis. Am J Bot. 91:65–72.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.