Abstract
Methods for detecting and identifying insects or insect fragments in food provides essential data to underpin regulatory responses aimed at protecting consumers. Complete mitochondrial genomes have demonstrated utility for high-resolution phylogenetic discrimination of closely related species; thus, high-quality references of complete mitochondrial genomes are valuable for applied detection and identification of insect species in food products.
Mitochondrial metagenomics (MMG) has been a highly successful approach for distinguishing a wide variety of species across a great breadth of biological taxonomy and provides a useful alternative to traditional polymerase chain reaction (PCR)-based methodologies (Gomez-Rodriguez et al. Citation2015; Linard et al. Citation2015; Pava-Ripoll et al. Citation2017). PCR primers often target a narrow range of insect species and do not always work equally well across different taxonomic groups. Advances in next-generation sequencing (NGS) platforms have minimized previous hurdles to acquisition of complete mitochondrial and plastid genomes, ushering in a new era of whole genome sequencing for eukaryotic genomes (Linard et al. Citation2015; Zhang et al. Citation2017).
Here, we present MitochonTrakr, a publicly accessible, high-quality reference collection of mitochondrial genomes from insect species that can potentially be found in food products. These reference genomes will facilitate target-independent sequence-based detection methodologies, providing sufficient information for discriminating closely related insect species, and creating new avenues for rapid detection and identification of insects in foods.
Fifty-two vouchered, authenticated reference insect species were retrieved from the USFDA-CFSAN insect collection. Insect species are from the orders Diptera (16), Coleoptera (18), Hymenoptera (8), Blattodea (7), Lepidoptera (2), and Orthoptera (1). Genomic DNA was extracted from whole or partial specimens using the DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA). NGS was performed using the NextSeq 550 System (Illumina, San Diego, CA) after normalizing the library to 1.8 pM. Sequence reads were concatenated and a reference-guided assembly was performed using NOVOPlasty (Dierckxsens et al. Citation2017) or MITObim 1.9.1 (Hahn et al. Citation2013). Contig sequences were annotated using MITOS (Bernt et al. Citation2013). Quality checks of annotations were performed as described by Cameron (Citation2014).
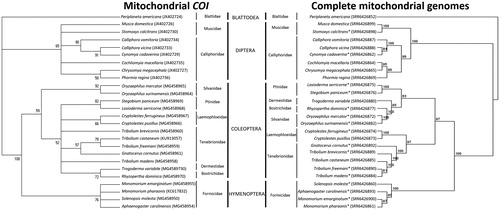
Phylogenetic analysis was performed using complete assembled mitochondrial genomes from 26 of the 52 insect species (). The maximum-likelihood (ML) tree with 100% bootstrap value supports the evolutionary relationship of the taxonomic families and orders of analysed species (Ishiwata et al. Citation2011; Sasaki et al. Citation2013).
Figure 1. Maximum-likelihood (ML) trees constructed using the barcode region of the mitochondrial cytochrome oxidase I (COI) gene (left) and using complete assembled and annotated mitochondrial genomes (right) of 26 insect species belonging to 10 families and four taxonomic orders. Number above each node indicates the bootstrap support. Branches with bootstrap values <50% are collapsed. The two phylogenetic trees demonstrate the utility of complete mitochondrial genomes for high resolution phylogenetic discrimination among taxa. Insect species have their corresponding accession numbers in parenthesis.*Newly annotated insect mitochondrial genomes.
This collection of insect mitochondrial genomes and their raw data are publicly available at the National Center for Biotechnology Information (NCBI) MitochonTrakr BioProject under accession number PRJNA423170, which is a component of the FDA MetaGenomeTrakr Project (PRJNA390622). Data currently comprise 52 authenticated insect specimen raw reads, 26 of which are assembled and annotated, including 12 newly annotated insect mitochondrial genomes.
Our high-quality reference collection of mitochondrial genomes from insect species was included in an in-house k-mer database (MITOkmerDB) that was developed for rapid k-mer-based identification of insect species. The database also includes 3407 publicly available insect mitochondrial genomes, covering 25 orders, and 7447 mitochondrial genomes from other eukaryotes including plants and fungi, among others. Signature k-mers were designed using non-overlapping sequence reads from our insect species. The number of sequence reads from insect species can be extracted from the total number of in silico matched reads using the tally of matches of the sequence data. This MITOkmerDB can be further used to detect and identify insect species found in complex food matrices.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Systematic Entomol. 39:400–411.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gomez-Rodriguez C, Crampton-Platt A, Timmermans M, Baselga A, Vogler AP. 2015. Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages. Methods Ecol Evol. 6:883–894.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:1–9.
- Ishiwata K, Sasaki G, Ogawa J, Miyata T, Su ZH. 2011. Phylogenetic relationships among insect orders based on three nuclear protein-coding gene sequences. Mol Phylogenetics Evol. 58:169–180.
- Linard B, Crampton-Platt A, Gillett C, Timmermans M, Vogler AP. 2015. Metagenome skimming of insect specimen pools: Potential for comparative genomics. Genome Biol Evol. 7:1474–1489.
- Pava-Ripoll M, Miller AK, Ziobro GC. 2017. Insect contaminants in foods: Detection limits of a qualitative PCR-based method. T4-09. IAFP Annual Meeting. July 9–12. 2017. Tampa, FL. 1.
- Sasaki G, Ishiwata K, Machida R, Miyata T, Su ZH. 2013. Molecular phylogenetic analyses support the monophyly of Hexapoda and suggest the paraphyly of Entognatha. Bmc Evolutionary Biology. 13:1–9.
- Zhang N, Ramachandran P, Wen J, Duke JA, Metzman H, McLaughlin W, Ottesen AR, Timme RE, Handy SM. 2017. Development of a reference standard library of chloroplast genome sequences, GenomeTrakrCP [Article]. Planta Med. 83:1420–1429.
