Abstract
We characterized the chloroplast (cp) genome sequence of Banara guianensis by using Illumina pair-end sequencing platform, and the complete whole cp genome was 158,577 bp. That contains a large single copy region (LSC) of 85,863 bp and a small single copy region (SSC) of 17,334 bp, which were separated by a pair of 27,690 bp inverted repeat regions (IRs). The cp genome has 112 annotated genes, including 76 protein-coding genes and 36 tRNA genes. The overall GC content of it is 36.5%, while the corresponding values of the LSC, SSC and IR regions are 34.3, 30.2 and 42.0%, respectively. Furthermore, the phylogenetic analysis suggested that the location of B. guianensis is a sister of A. serrata and is an outgroup of other Salicaceae species.
Banara, having ca. 36 species distributed in South America, is a genus in family Salicaceae which formerly in family Flacourtiaceae (The plant list Citation2018). Due to the lack of genome data, the phylogenetic placement of Banara and the evolutionary relationship with other Salicaceae genera is not clear. In this study, we used illumina pair-end sequencing data to assemble and characterize the complete chloroplast genome sequence of Banara guianensis and to uncover the phylogenetic position in family Salicaceae. The voucher specimen was collected at Napo, Ecuador (1.392S, 77.695W) and stored at Gray Herbarium of Harvard University (Mac H. Alford 3089).
The total genomic DNA was extracted from dry leaves using a modified CTAB method (Doyle and Doyle Citation1987). And the filtered reads were assembled by the program named NOVOPlasty (Dierckxsens et al. Citation2017) and used the sequenced chloroplast genome Abatia parviflora (Unpublished) as the reference. Then, we annotated the assembled chloroplast genome by Plann (Huang and Cronk Citation2015), and the annotation was corrected using Geneious (Kearse et al. Citation2012). In addition, the physical map of the chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013). The complete chloroplast genome together with gene annotations was submitted to GenBank under the accession number of MH937752. To further investigate its phylogenetic placement, we construct a neighbour-joining tree (Saitou and Nei Citation1987) using the complete chloroplast genome sequences data of 16 seed plants. We were first aligned these 16 chloroplast genome sequences using MAFFT (Katoh and Standley Citation2013), and the phylogenetic tree was then constructed using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates.
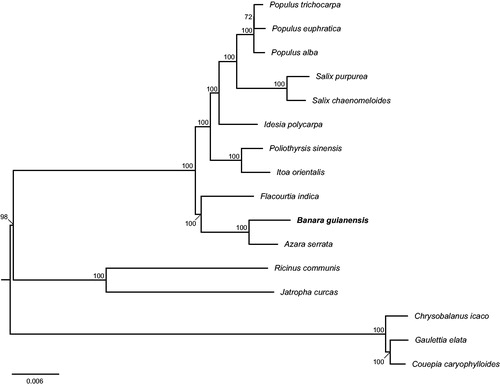
Our assembled chloroplast genome of B. guianensis is 158,577 base pairs (bp) in length, containing a large single-copy (LSC) region of 85,863 bp, a small single-copy (SSC) region of 17,334 bp, and two inverted repeat (IR) regions of 27,670 bp. In addition, the chloroplast genome has 112 genes in total, including 76 protein-coding genes and 36 tRNA genes. Most of them are in a single copy; however, seven protein-coding genes (i.e. ndhB, rpl2, rpl23, rps12, rps19, rps7 and ycf2), and seven tRNA genes (i.e. trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) occur in double copies. The overall GC-content of the whole cp is 36.5%, while the corresponding values of the LSC, SSC, and IR regions are 34.3, 30.2 and 42.0%, respectively. Furthermore, the phylogenetic placement of B. guianensis is a sister of A. serrata and is an outgroup of other Saliaceae species with 100% bootstrap support (). The location of other species consisted with the previous study (Zhang et al. Citation2018) of Salicaceae phylogenetic analysis.
Figure 1. Phylogenetic relationships of 16 seed plants based on plastome sequences. Bootstrap percentages are indicated for each branch. GenBank accession numbers: Azara serrata (MH719101), Chrysobalanus icaco (KJ414480), Couepia caryophylloides (KX180053), Flacourtia indica (MG262341), Gaulettia elata (KX180066), Idesia polycarpa (KX229742), Itoa orientalis (MG262342), Jatropha curcas (FJ695500), Poliothyrsis sinensis (MG262343), Populus alba (AP008956), Populus euphratica (KJ624919), Populus trichocarpa (EF489041), Ricinus communis (JF937588), Salix chaenomeloides (MG262362) and Salix purpurea (KP019639).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol Biol Evol. 33:1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41(Web Server issue): W575–W581.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- The Plant List. 2018. Version 1.1. Published on the Internet; (accessed 2018 September 20). http://www.theplantlist.org/
- Zhang L, Xi Z, Wang M, Guo X, Ma T. 2018. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol Evol.8: 7817–7823.
