Abstract
The complete mitochondrial genome of an oleaginous microalga Vischeria stellata strain SAG 33.83 was sequenced and annotated. The circular genome was 42,608 bp long with 26.7% GC content. The mitochondrial genome of V. stellata strain SAG 33.83 contained 36 protein-coding genes (PCGs), 27 transfer RNA genes (tRNAs), three ribosomal RNA genes (rRNAs). Furthermore, the phylogenetic tree was constructed to validate the evolutionary relationship based on the complete mitogenomes of V. stellata strain SAG 33.83 through combining with seven species of Eustigmatophyceae, two species of Phaeophyceae and one species of Synurophyceae algae. Phylogenetic analysis revealed that Eustigmatophyceae were more closely related to Synurophyceae than Phaeophyceae and V. stellata strain SAG 33.83 had close genetic relationship with Vischeria sp. CAUP Q 202.
Vischeria stellata strain SAG 33.83, an edaphic unicellular microalga, classified into family Eustigmatophyceae, was considered as a promising oleaginous microalga for the production of biofuels and bioproducts (Gao et al. Citation2016; Wang et al. Citation2018). It can accumulate large amounts of lipids (66.8% of dry weight (DW)) comprising two important nutritional fatty acids, palmitoleic acid (28% of DW) and eicosapentaenoic acid (1.5% of DW), and other fatty acids. In addition, it could store certain amount of β-carotene (1–2% of DW), which was considered to be a potential nutraceutical. The previous researches about V. stellata focused on cytological character, culture condition and lipid metabolism (Gao et al. Citation2016). The phylogenetic information of V. stellata, such as 18S rDNA and rbcL gene, is limited.
According to endosymbiosis theory, mitochondrial genome was organelle genetic substance of the eukaryotic cell and directly descendant from endosymbiont host (heterotrophic eukaryotes) and could provide phylogenetic information of endosymbiotic origins (Lang and Burger Citation2012). Nevertheless, it was not clear whether the mitochondrial genome of V. stellata plays an important role in lipids metabolism. To reveal the gene content and structure of mitochondria, the whole-genome of V. stellate strain SAG 33.83 was sequenced through Illumina sequencing technology, and the complete mitochondrial genome was obtained.
The origin sample of V. stellata strain SAG 33.83 was collected from Isle Lavsa, Dalmatia, Europe (43.7521° N, 15.3695° W) and deposited in the Culture Collection of Algae at Gottingen University. The total genomic DNA of V. stellata strain SAG 33.83 was extracted by MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa, China) and stored in our laboratory. Then, the purified DNA was used to build fractionated genomic libraries and sequenced with Illumina HiSeq4000 platform by BGI Biotechnology Co. Ltd (Shenzhen, China). After the quality filtration of raw reads, the high-quality reads were assembled by SPAdes 3.9.0 (Bankevich et al. Citation2012). One contig of the mitochondrial DNA was identified by comparing the sequence similarity to other eustigmatophycean mitochondrial genomes. Finally, the mitochondrial genome annotation was performed by the UGENE ORFs finder, tRNAscan-SE and RNAmmer 1.2 Server.
The complete mitochondrial genome of V. stellata strain SAG 33.83 was submitted to NCBI (GenBank accession number: MH981596). The length of the mitochondrial genome was 42,608 bp of circular genome, with 26.56% GC content. It contained 3 rRNA genes for 5S, small subunit and large subunits ribosomal RNA (rRNA), 27 tRNA genes and 36 protein-coding genes include four atp genes, three cox genes, 10 nad genes, 16 ribosomal protein genes, one apocytochrome b, one tatC genes and one open reading frames (ORF405).
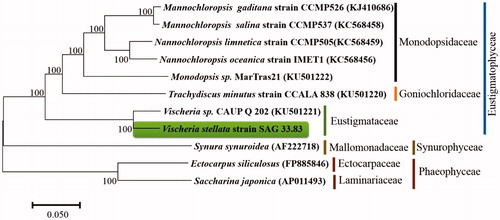
Phylogenetic analysis was performed by 11 complete mitogenomes from seven species of Eustigmatophyceae (Mannochloropsis gaditana strain CCMP526 (GenBank accession number:KJ410686), Mannochloropsis salina strain CCMP537 (KC568458), Nannochloropsis limnetica strain CCMP505 (KC568459), Nannochloropsis oceanica strain IMET1 (KC568456), Monodopsis sp. MarTras21 (KU501222), Trachydiscus minutus strain CCALA 838 (KU501220), Vischeria sp. CAUP Q 202 (KU501221)) (Wei et al. Citation2013; Ševčíková et al. Citation2016), two species of Phaeophyceae (Ectocarpus siliculosus (FP885846), Saccharina japonica (AP011493) (Cock et al. Citation2010) and one species of Synurophyceae (Synura synuroidea (AF222718)) (Chesnick et al. Citation2000) and V. stellata strain SAG 33.83 in this study. The concatenated alignments of 11 algal mitochondrial genomes were calculated by HomBlocks (Bi et al. Citation2017), resulting in alignments of each species with 11,151 bp, including cob, cox1, cox3, nad1, nad4, nad5, nad7, small subunit and large subunits rRNA. The phylogenetic tree was constructed based on neighbour-joining method by MEGA7 (Kumar et al. Citation2016), relied on Gamma distribution model with 1000 bootstrap replicates. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The phylogenetic analysis results supported that the mitochondrial genes of Eustigmatophyceae were more closely related to Synurophyceae than Phaeophyceae (). Meanwhile, the phylogenetic tree showed that the V. stellata strain SAG 33.83 was clustered in the Eustigmataceae clade and genetically close to Vischeria sp. CAUP Q 202 (KU501221) (Ševčíková et al. Citation2016).
Declaration statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 465:617.
- Chesnick JM, Goff M, Graham J, Ocampo C, Lang BF, Seif E, Burger G. 2000. The mitochondrial genome of the stramenopile alga Chrysodidymus synuroideus. Complete sequence, gene content and genome organization. Nucleic Acids Res. 28:2512–2518.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2017. Homblocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Gao B, Yang J, Lei X, Xia S, Li A, Zhang C. 2016. Characterization of cell structural change, growth, lipid accumulation, and pigment profile of a novel oleaginous microalga, Vischeria stellata (Eustigmatophyceae), cultured with different initial nitrate supplies. J Appl Phycol. 28:821–830.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lang BF, Burger G. 2012. Mitochondrial and eukaryotic origins: a critical review. Adv Botanical Res. 63:1–20.
- Ševčíková T, Klimeš V, Zbránková V, Strnad H, Hroudová M, Vlček C, Eliáš M. 2016. A comparative analysis of mitochondrial genomes in eustigmatophyte algae. Genome Biol Evol. 8:705–722.
- Wang F, Gao B, Huang L, Su M, Dai C, Zhang C. 2018. Evaluation of oleaginous eustigmatophycean microalgae as potential biorefinery feedstock for the production of palmitoleic acid and biodiesel. Bioresource Technol. 270:30–37.
- Wei L, Xin Y, Wang D, Jing X, Zhou Q, Su X, Jia J, Kang N, Chen F, Hu Q, Xu J. 2013. Nannochloropsis plastid and mitochondrial phylogenomes reveal organelle diversification mechanism and intragenus phylotyping strategy in microalgae. BMC Genomics. 14:534.