Abstract
Lack of mitochondrial genome data of Scleractinia is hampering progress across genetic, systematic, phylogenetic and evolutionary studies. In this study, the complete mitogenome sequence of the stony coral, Acropora valida (Dana, 1846), has been decoded for the first time by next-generation sequencing and genome assembly. The assembled mitogenome was 18,385 bp in length, containing 13 protein-coding genes (PCGs), two transfer RNAs and two ribosomal RNAs. All genes were encoded on the same strand. The A + T content of the mitochondrial genome was 62.0% (25.0% A, 37.0% T, 24.3% G, and 13.8% for C). The complete mitogenome of Acropora valida has the same gene content and structure as other Acropora species. The complete mitogenome provides essential and important DNA molecular data for further phylogenetic and evolutionary analyses for coral phylogeny.
Reef-building coral species of the order Scleractinia play an important role in shallow tropical seas by providing an environmental base for the ecosystem (Fukami et al. Citation2000). While traditional morphology-based systematics cannot clearly reflect all the evolutionary relationships of Scleractinia, molecular data have therefore become increasingly important in recent years to overcome the limitations of morphological analyses among scleractinians (Arrigoni et al. Citation2017; Terraneo et al. Citation2017). The unique characters of mitochondrial genome DNA (mitogenome), which include small size, fast evolutionary rate, simple structure, maternal inheritance and high informational content, suggest that the constituting loci could be powerful markers for resolving ancient phylogenetic relationships (Boore Citation1999; Sun et al. Citation2003; Geng et al. Citation2016). However, the complete mitochondrial genomes of stony corals that we can find in NCBI (National Centre for Biotechnology Information) are less than 100 species. Acropora valida (Dana, 1846), one of the important members of Acroporidae, is widely distributed from Indian Ocean to eastern Pacific Ocean. In the present study, we determined the complete mitogenome sequence of A. valida to provide basic information for further evolutionary and phylogenetic analyses.
The specimen (voucher no. DYW8) of A. valida was collected from Daya Bay in Guangdong, China. Total genomic DNA was extracted using the DNeasy tissue Kit (Qiagen China, Shanghai) and kept at 4°°C for subsequent use. We used next-generation sequencing to perform low-coverage whole-genome sequencing according to the protocol (Tian and Niu Citation2017). Initially, the raw next-generation sequencing reads generated from HiSeq 2000 (Illumina, San Diego, CA). About 0.17% raw reads (6522 out of 3,897,136) were de novo assembly by using commercial software (Geneious V9, Auckland, New Zealand) to produce a single, circular form of complete mitogenome with about an average 74 × coverage.
The complete mitogenome of A. valida was 18,385 bp in size (GenBank accession number: MH141598) and its overall base composition was 25.0% for A, 37.0% for T, 24.3% for G, and 13.8% for C. The protein coding, rRNA and tRNA genes of A. valida mitogenome were predicted by using DOGMA (Wyman et al. Citation2004), ARWEN (Laslett and Canback Citation2008), MITOS (Bernt et al. Citation2013) tools and manually inspected. The complete mitogenome of A. valida included unique 13 protein-coding genes (PCGs), two transfer RNA genes (tRNAMet, tRNATrp) and two ribosomal RNA genes. All PCGs, tRNA and rRNA genes were encoded on H-strand. Protein coding genes preferred base T, tRNA genes preferred base G and rRNA genes preferred base A.
The PCGs was 11,866 bp in size, and its base composition was 22.2% for A, 13.9% for C, 23.7% for G and 40.2% for T. The PCG of ND5 had a 11,976 bp intron insertion. It was important to note that six of 13 PCGs started with GTG codon, seven of 13 PCGs started with ATG codon, only ND6 gene started with ATA codon. Five PCGs were inferred to terminate with TAA (ND1, ND2, ND6, ND4L and COI), other PCGs with TAG (Cyt b, ATP6, ND4, ND3, ND5, COII, COIII and ATP8). Among 13 PCGs, the longest one was ND5 gene (1,836 bp), whereas the shortest was ATP8 gene (219 bp). There were 19 bp overlapping nucleotides between ATP8 and COI, and the number of non-coding nucleotides between different genes varied from 30 to 1127 bp.
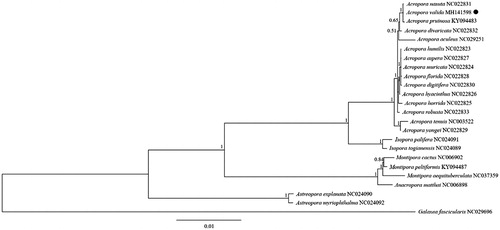
To validate the phylogenetic position of A. valida within the Scleractinia, the complete mitogenome sequences of 22 Acroporidae species downloaded from NCBI were incorporated together for phylogenetic analysis. Galaxea fascicularis (Linnaeus, 1767) belonging to Euphylliidae was used as outgroup for tree rooting. The phylogenetic trees were built using Bayesian inference (BI) analysis by Mrbayes 3.12 (Huelsenbeck and Ronquist Citation2001) based on 13 PCGs binding sequence. The substitution model selection was conducted by a comparison of Akaike Information Criterion (AIC) scores with jModelTest 2 (Darriba et al. Citation2012). GTR + G model was chosen as the best-fitting model for BI analyses and the node reliability was estimated. Result showed that A. valida was closely related to Acropora nasuta with high bootstrap value supported (). In conclusion, the complete mitogenome of the A. valida deduced in this study provides essential and important DNA molecular data for further phylogenetic and evolutionary analyses for stony coral phylogeny.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Arrigoni R, Berumen ML, Huang DW, Terraneo TI, Benzoni F. 2017. Cyphastrea (Cnidaria: Scleractinia: Merulinidae) in the Red Sea: phylogeny and a new reef coral species. Invert Systematics. 31:141–156.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Fukami H, Omori M, Hatta M. 2000. Phylogenetic relationships in the coral family acroporidae, reassessed by inference from mitochondrial genes. Zool Sci. 17:689–696.
- Geng X, Cheng R, Xiang T, Deng B, Wang Y, Deng D, Zhang H. 2016. The complete mitochondrial genome of the Chinese Daphnia pulex (Cladocera, Daphniidae). Zookeys. 27:47–60.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Terraneo TI, Arrigoni R, Benzoni F, Tietbohl MB, Berumen ML. 2017. Exploring the genetic diversity of shallow-water Agariciidae (Cnidaria: Anthozoa) from the Saudi Arabian Red Sea. Mar Biodivers. 47:1065–1078.
- Tian P, Niu WT. 2017. The complete mitochondrial genome of the Acropora pruinosa. Mitochondr DNA Part B. 2:652–653.
- Sun H, Zhou K, Song D. 2003. Mitochondrial genome and phylogenetic reconstruction of arthropods. Zool Res. 24:467–479.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.