Abstract
Pericallis hybrida is a very prevalent decorative plant worldwide. Here, we report the assembly of the complete nucleotide sequence of the P. hybrida chloroplast genome using the program MITObim v1.7 after whole genome shotgun sequencing. The complete chloroplast genome of P. hybrida (GenBank accession number: KT285537) consists of 151,267 bp and includes a pair of inverted repeats (IR) of 23,595 bp separated by one small and one large single copy region (SSC and LSC) of 18,330 bp and 85,747 bp, respectively. The genome has a 37.3% GC content and contains 132 genes coding for 87 proteins. Phylogenetic analyses using complete chloroplast genome revealed that P. hybrida belongs to the Senecioneae of Asteraceae family, which is conformed to the traditional classification.
Pericallis hybrida, as a perennial herb, is a species of Pericallis which belongs to the Senecioneae of the highly diverse Asteraceae family. However, so far, in genus Pericallis, no complete chloroplast genome has been sequenced and assembled. The complete chloroplast genome of Jacobaea vulgaris from the Senecioneae of the Asteraceae family is the only one that has been released on the GenBank of NCBI database (Li et al. Citation2015). In this study, the complete chloroplast genome of P. hybrida was presented, to contribute to furnish resourced organelle genome of Plantae, which will be helpful for species identification, phylogenetic relationships, and plant classification.
The sample for DNA extraction was obtained from the fresh leaves of P. hybrida in the greenhouse of National Engineering laboratory for Resource Developing of Endangered Chinese Crude Drugs in Northwest of China (108°53′30″E, 34°9′14 ″N). Total genomic DNA was extracting and whole-genome sequencing was conducted by Shanghai Genesky Biotechnologies Inc. (Shanghai, China) with an Illumina Hiseq 2500 Sequencing System (Illumina, San Diego, CA). A total of 10.61 M raw reads, having an average fragment length of 126 bp were retrieved. And then, those raw rends were quality trimmed with default parameters using CLC Genomics Workbenchv7.5 (Patel and Jain Citation2012). After trimming, there were 10.54 M trimmed reads, having an average fragment length of 122 bp, which were then used for the chloroplast genome reconstruction. The complete chloroplast genome was assembled using the program MITObimv1.7 (Christoph et al. Citation2013) with the J. vulgaris (GenBank: NC_015543) genome as the initial reference. The assembled sequence was annotated using GENEIOUS R8 (Kearse et al. Citation2012) by comparing it with the complete chloroplast genomes of J. vulgaris. At last, the annotated chloroplast genome sequence was submitted to GenBank with the accession number KT285537.
The P. hybrida complete circular chloroplast genome is 151,267 bp in size and contains a pair of inverted repeat (IR) regions of 23,595 bp each, a large single-copy (LSC) region of 85,747 bp, and a small single-copy (SSC) region of 18,330 bp. The chloroplast genome contains 132 genes (109 species) including 87 protein-coding genes (77 species), with 17 of these genes being duplicated in the IR regions, eight ribosomal RNA genes (four species), and 37 transfer RNA genes (28 species). Fifteen genes have introns ranging from 153 to 2545 bp in length. There are 13 genes containing only a single intron. Additionally, there are two genes, ycf3 and clpP, that contain two introns. In the chloroplast genome, 66.4% was the protein-coding region. In addition, the nucleotide composition is asymmetrical (31.2% A, 18.4% C, 18.9% G and 31.5% T) with an overall GC content of 37.3%.
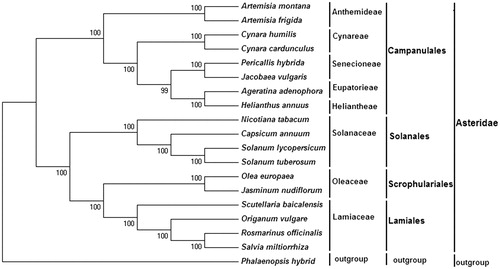
The complete chloroplast genome of P. hybrid and other species from Asteraceae were used to construct a phylogenetic tree. A neighbour-joining (NJ) tree was performed with Mega 6.0 using 1000 bootstrap replicates (Tamura et al. Citation2013). The result shows P. hybrida belongs to the Senecioneae of Asteraceae family, which is conformed to the traditional classification ().
Figure 1. Phylogenetic tree of 18 Asteridae species, based on the complete chloroplast genome. The tree was generated by neighbor-joining (NJ) method using the MEGA6 program. Bootstrap values are indicated on the branches. The tribe names of the corresponding species are indicated on the right margin. Accession numbers: Solanum tuberosum DQ386163, Jasminum nudiflorum DQ673255, Olea europaea GU931818, Origanum vulgare JX880022, Capsicum annuum KJ619462, Cynara cardunculus KM035764, Solanum lycopersicum KP331414, Scutellaria baicalensis KR233163, Helianthus annuus NC_007977, Jacobaea vulgaris NC_015543, Ageratina adenophora NC_015621, Salvia miltiorrhiza NC_020431, Artemisia frigida NC_020607, Artemisia Montana NC_025910, Cynara humilis NC_027113, Rosmarinus officinalis NC_027259, Nicotiana tabacum Z00044, Phalaenopsis hybrid NC_025593 and Pericallis hybrida KT285537.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Christoph H, Lutz B, Bastien C. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev. 90:157.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 44:2725.
