Abstract
Crossostephium chinense is a common traditional Chinese medicinal plant used to cure arthralgia and rheumatism. The previous studies mainly focussed on its chemical compositions, with the genetic background and resources remaining scarce. Here, we conducted genome skimming for C. chinense, and using these data we not only assembled its complete chloroplast genome, but also reconstructed the phylogenetic relationship between C. chinense and Artemisia. The cp genomes of two C. chinense individuals ranged from 151,024 bp to 151,097 bp in length and had a typical quadripartite structure with a conserved genome arrangement. The cp genome encodes 114 unique genes, consisting of 80 protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Phylogenetic analysis indicates that C. chinense is sister to Artemisia annua and Artemisia fukudo.
Crossostephium Less. (Asteraceae) is a monotypic genus which contains a single species, C. chinense (L.) Makino. Crossostephium chinense is a traditional Chinese medicinal plant recorded in many Chinese herbal books and has been used as a folk herbal drug in South China (Wu et al. Citation2009). The whole plants of C. chinense are used to treat furuncle, carbuncle, diabetes, and cold. Previous studies of C. chinense mainly focussed on its phytochemical composition (Yang et al. Citation2008). However, researches regarding its genetic background are extremely scarce, which will hinder its effective utilities and protection.
In this study, two individuals of C. chinense were selected in Zhejiang (China; 121°09′28.44″E, 27°49′32.16″N) and Motobu (Japan; 127°53′48.48″E, 26°39′28.80″N) for genome skimming. Genomic DNA was extracted from silica-dried leaf tissue using Plant DNAzol Reagent (LifeFeng, Shanghai, China) according to the manufacturer’s protocol. Voucher specimens (Pan Li LP161101-2: ZJWZ and Pan Li LP173485-1: JPBB) were deposited at the Herbarium of Zhejiang University (HZU). The high-quality DNA was sheared (yielding ≤800 bp fragments) and the short-insert (500 bp) paired-end libraries preparation and sequencing were performed on Illumina HiSeq 2500 by Beijing Genomics Institute (Wuhan, China). The raw data were filtered by quality with Phred score <30 and the remaining sequences were assembled into contigs using the CLC de novo assembler beta 4.06 (CLC Inc., Rarhus, Denmark) to reconstruct the cp genome with Artemisia frigida (GenBank accession number: NC_020607) as a reference. The annotation of the cp genome was performed through the online program Dual Organellar Genome Annotator (Wyman et al. Citation2004), and the circular gene maps were drawn by the OrganellarGenomeDRAW tool (OGDRAW) following by manual modification (Lohse et al. Citation2013). The phylogenetic tree was reconstructed based on whole cp genome sequences of nine Artemisia species and two Crossostephium individuals using the maximum likelihood (ML) method, with two Chrysanthemum species as outgroup. The ML analysis was implemented in RAxML-HPC v8.1.11 on the CIPRES cluster (Miller et al. Citation2010).
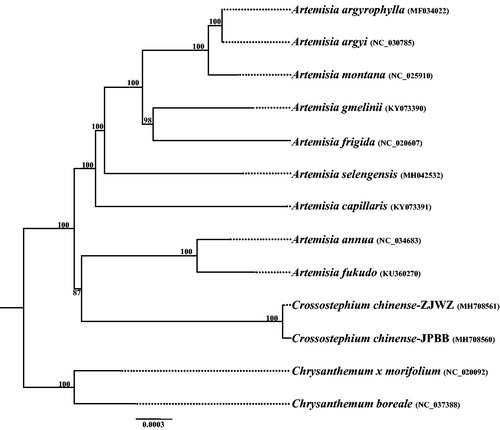
The complete chloroplast genomes of the two C. chinense individuals are 151,024 bp and 151,097 bp, respectively. Both chloroplast genomes shared the common feature of comprising two copies of IR (both are 24,975 bp) separated by the LSC (82,786 bp and 82,819 bp) and SSC (18,288 bp and 18,328 bp) regions. The overall GC content was 37.40% for the two C. chinense individuals, whereas the GC content in the LSC, SSC, and IR regions were 35.50, 30.70, and 43.10%, respectively. The two chloroplast genomes encoded an identical set of 133 genes, of which 114 were unique and 19 were duplicated in the IR regions. Among the 114 unique genes, 80 are protein-coding genes, 30 are tRNA genes, and four are rRNA genes. The cp genome sequences were registered in GenBank with the accession number MH708561 and MH708560. The phylogeny revealed that the Artemisia species are paraphyletic, which is consistent with the previous study (Watson et al. Citation2002), and the two C. chinense individuals are sisters to A. annua and A. fukudo with moderate value support ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575.
- Miller M-A, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Comput Environ Workshop (GCE), 2010. IEEE. :1–8.
- Watson L-E, Bates P-L, Evans T-M, Unwin M-M, Estes J-R. 2002. Molecular phylogeny of Subtribe Artemisiinae (Asteraceae), including Artemisia and its allied and segregate genera. BMC Evol Biol. 2:17.
- Wu Q, Zou L, Yang X-W, Fu D-X. 2009. Novel sesquiterpene and coumarin constituents from the whole herbs of Crossostephium chinense. J Asian Natural Prod Res. 11:85.
- Wyman S-K, Jansen R-K, Boore J-L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang X, Zou W-L, Wu Q, Fu D-X. 2008. Studies on chemical constituents from whole plants of Crossostephium chinense. Zhongguo Zhong Yao Za Zhi. 33:905–908 (In Chinese).