Abstract
We sequenced complete mitochondrial genome of upland rice Oryza sativa (Liliopsida: Poales: Poaceae) with length of 444,883 bp. The genome has similar codon usage and gene organization to those of mitogenome reported for other Poales. It includes 55 protein-coding genes, 6 ribosomal RNA genes, and 26 transfer RNA genes. The overall base composition is A (27.94%), T (28.46%), G (21.86%), C (21.74%), so the relatively balanced base content was detected. Phylogenetic analyses showed that O. sativa is closely related to Oryza rufipogon that is also a member of the same genus, which is a widely accepted view.
In this study, we presented the mitochondrial genome of the Oryza species, Oryza sativa, which belong to Oryzeae. Oryzeae (Poaceae) is a tribe belonging to Poaceae and consists of about 12 genera. This tribe contains many economically important species and is distributed in tropical and temperate regions worldwide. Recently, investigation on the genus Oryza and its related genera in the Oryzeae has attracted much attention for their important economic value and taxonomic position in the study of plant biology in general (Duistermaat Citation1987; Gaut Citation2002). Specimen in this study was collected in 2007 from Yuanyang County, Honghe Autonomous Prefecture, Yunnan Province, southwest China. The rice seeds were stored at −25ºC in Tianjin Crop (rice) Research Institute, Tianjin Academy of Agricultural Sciences until used in this experiment.
The entire mitochondrial genome of O. sativa was 444,883 bp and deposited in the GenBank under the accession number MH665664 in NCBI. The genome has similar codon usage and gene organization to those of mitogenome reported for other Poaceae. Genetic DNA from rice seedling was individually extracted. Annotation of the complete assembled mitochondrial genome was performed with a Dual Organellar GenoMe Annotator (DOGMA, Jansen Lab, Austin) (Wyman et al. Citation2004). The total base composition was A (27.94%), T (28.46%), G (21.86%), C (21.74%), so the relatively balanced base content was detected.
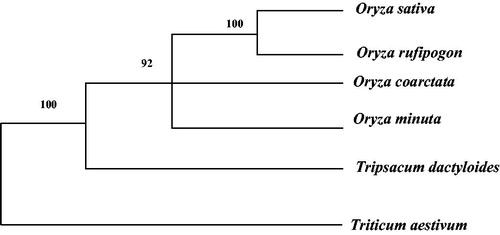
The phylogenetic analyses in this study concur with the proposed and widely accepted view of the internal relationships of Oryzeae, O. sativa and O. rufipogon are sister taxa and O. minuta are more original (Kellogg and Watson Citation1993; Watson and Dallwitz Citation1999; Kellogg Citation2000; Ge et al. Citation2002). The complete mitochondrial genomes of five other species are available from GenBank and their scientific names and accession numbers are as follows: Oryza rufipogon (AP012527), Oryza coarctata (MG429050), Oryza minuta (KU176938), Tripsacum dactyloides (NC_008362), Triticum aestivum (NC_036024). The consensus tree topology showed Oryzeae to be completely monophyletic taxa () and the relation between tribes was observed having 1000 bootstrap value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Duistermaat H. 1987. A revision of Oryza (Gramineae) in Malesia and Australia. Blumea. 32:157–193.
- Gaut BS. 2002. Evolutionary dynamics of grass genomes. New Phytologist. 154:15–28.
- Ge S, Li A, Lu B-R, Zhang S-Z, Hong D-Y. 2002. A phylogeny of the rice tribe Oryzeae (Poaceae) based on matK sequence data. Am J Botany. 89:1967–1972.
- Kellogg EA. 2000. The grasses: a case study in macroevolution. Ann Rev Ecol Systemat. 31:217–238.
- Kellogg EA, Watson L. 1993. Phylogenetic studies of a large data set. I. Bambusoideae, Andropogonodae, and Pooideae (Gramineae). Botanical Rev. 59:273–343.
- Watson L, Dallwitz MJ. 1999. Grass genera of the world: descriptions, illustrations, identification, and information retrieval; including synonyms, morphology, anatomy, physiology, phytochemistry, cytology, classification, pathogens, world and local distribution, and references; [accessed 1999 Aug 18]. http://biodiversity.uno.edu/delta/
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.