Abstract
In the present report, we describe the complete mitochondrial genome of Psolodesmus mandarinus, from the Hualien County, Taiwan. This mitogenome is 15,176 bp long, containing 13 protein-coding, 22 tRNA, and two rDNA genes. Nucleotide composition of the whole mitogenome was 41% for A, 25.92% for T, 19.06% for C, and 14.03% for G. The AT and GC skewness of mitogenome sequence was 0.225 and 0.152, showing the A-skew and C-skew. The reconstructed phylogenetic relationships of 23 Odonata species based on 13 protein-coding genes were highly supported. P. mandarinus grouped within the clade including the other three Calopterygidae genera, Mnais, Vestalis, and Atrocalopteryx was solid supported (100%). It was relatively close to Mnais costalis in the phylogenetic analysis. Our study will be useful for the population genetics, biogeography, and conservation of P. mandarinus in the future.
There are only two species in the genus Psolodesmus McLachlan: Psolodesmus mandarinus in Taiwan and P. kuroiwae in Yaeyama islands in the Japanese Ryukyu Archipelago (Hämäläinen Citation2004; Karjalainen and Hämäläinen Citation2013). P. mandarinus is distributed in the streams up to 1500 m in Taiwan. The flight period is from March to December. The larval habitat is forest streams. Adults appear near water in the forest. Males often perch near females for courtship. Females usually oviposit on moss-covered stones, submerged leaves, and aquatic plants (Wang Citation2000). This is the first report of complete mitochondrial sequences for the genus Psolodesmus.
The specimen of Psolodesmus mandarinus, in this study, collected from Fuyuan, Hualien County, Taiwan in November 2015. Total genomic DNA was extracted from the adult’s thorax using the QuickExtract™ DNA Extraction Solution kit (Epicentre, Madison, WI) following the supplier’s instructions. The voucher specimen’s genome DNA and partial specimen were deposited in the Taiwan Forestry Research Institute, Taipei, Taiwan. The complete mitogenome of P. mandarinus was sequenced using the next-generation sequencing method (Illumina MiSeq, San Diego, CA). A total of 2.2 Gb next-generation sequencing paired-end reads were used to assemble the complete mitogenome sequence. The CLC Genomics Workbench (QIAGEN) was used for sequence quality analysis, data trimming, and de novo assembling. The locations of the protein-coding genes, ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) were predicted by using MITOS Web Server (Bernt et al. Citation2013) and identified by alignment with other mitogenomes of Odonata. The AT and CG skew was calculated according to the following formulas: AT skew = (A – T)/(A + T) and GC skew = (C – G)/(C + G) (Perna and Kocher Citation1995). The phylogenetic analysis was done by using MEGA6 (Tamura et al. Citation2013). The complete mitogenome of P. mandarinus is 15,176 bp in length (GenBank Accession No. MF150044), including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and one control region. The total nucleotide composition of the P. mandarinus mitogenome was 41% for A, 25.92% for T, 19.06% for C, and 14.03% for G. The AT and GC skewness of mitogenome sequence was 0.225 and 0.152, showing the A-skew and C-skew. The skew statistic of protein coding gene was 0.248 and 0.147, showing the same A-skew and C-skew. The gene rearrangement of the P. mandarinus mitogenome is identical to the ancestral insect type (Cameron Citation2014).
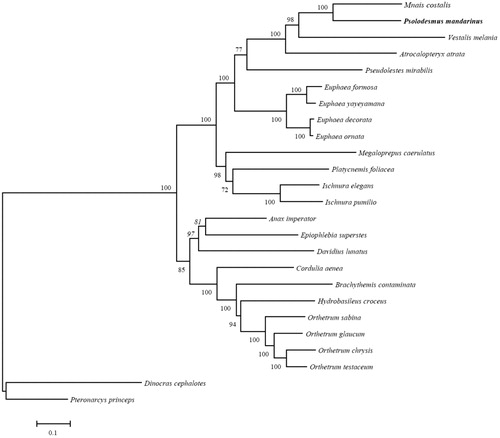
We reconstructed the phylogenetic relationships of 23 Odonata species and two species of outgroup (Pteronarcys princeps and Dinocras cephalotes) based on 13 protein-coding genes with Maximum Likelihood (ML) criteria by using MEGA6. Bootstrap values (1000 replications) greater than 70% are shown at the branch nodes (). The majority of nodes had support values higher than 70% and 14 were 100% supported. P. mandarinus grouped within the clade including the other three Calopterygidae genera, Mnais, Vestalis, and Atrocalopteryx was solid supported (100%). P. mandarinus was relatively close to Mnais costalis in the phylogenetic analysis, consistent with the result of Dumont et al. (Citation2005). Mitochondrial markers from our study will be useful for the population genetics, biogeography, and conservation of P. mandarinus in the future.
Figure 1. Phylogenetic tree of the 23 Odonata species based on the sequence of 13 protein-coding genes. The tree was reconstructed with the Maximum Likelihood (ML) criteria using MEGA v.6 (Tamura et al. Citation2013). Bootstrap values (1000 replications) greater than 70% are shown at the branch nodes. Anax imperator (NC031821) (Herzog et al. Citation2016), Atrocalopteryx atrata (NC027181), Brachythemis contaminate (NC026305) (Yu et al. Citation2016), Cordulia aenea (JX963627), Davidius lunatus (NC012644) (Lee et al. Citation2009), Epiophlebia superstes (NC023232) (Wang et al. Citation2015), Euphaea decorate (KF718294), Euphaea formosa (NC014493) (Lin et al. Citation2010), Euphaea ornate (KF718295), Euphaea yayeyamana (KF718293), Hydrobasileus croceus (NC025758), Ischnura elegans (NC031824) (Feindt et al. Citation2016a), Ischnura pumilio (KC878732) (Lorenzo-Carballa et al. Citation2014), Megaloprepus caerulatus (NC031823) (Feindt et al. Citation2016b), Mnais costalis (AP017642), Orthetrum chrysis (KU361233) (Yong et al. Citation2016), Orthetrum glaucum (KU361232) (Yong et al. Citation2016), Orthetrum sabina (KU361234) (Yong et al. Citation2016), Orthetrum testaceum (KU361235) (Yong et al. Citation2016), Platycnemis foliacea (NC027180), Pseudolestes mirabilis (NC020636), Psolodesmus mandarinus (in this study), Vestalis melania (NC023233) (Chen et al. Citation2015), Pteronarcys princeps (AY687866) (Stewart and Beckenbach Citation2006), Dinocras cephalotes (KF484757).
Acknowledgements
We are grateful to Chin-Hua Ai for his field work and Yen-Wei Chou for his assistance in DNA data downloading.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Chen MY, Chaw SM, Wang JF, Villanueva RJ, Nuñeza OM, Lin CP. 2015. Mitochondrial genome of a flashwing demoiselle, Vestalis melania from the Philippine Archipelago. Mitochondrial DNA. 26:720–721.
- Dumont HJ, Vanfleteren JR, De Jonckheere JF, H Weekers PH. 2005. Phylogenetic relationships, divergence time estimation, and global biogeographic patterns of calopterygoid damselflies (Odonata, Zygoptera) inferred from ribosomal DNA sequences. Syst Biol. 54:347–362.
- Feindt W, Herzog R, Osigus HJ, Schierwater B, Hadrys H. 2016a. Short read sequencing assembly revealed the complete mitochondrial genome of Ischnura elegans Vander Linden, 1820 (Odonata: Zygoptera). Mitochondrial DNA B Resour. 1:574–576.
- Feindt W, Osigus HJ, Herzog R, Mason CE, Hadrys H. 2016b. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA B Resour. 1:497–499.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hämäläinen M. 2004. Caloptera damselflies from Fujian (China), with description of a new species and taxonomic notes (Zygoptera: Calopterygoidea). Odonatologica. 33:371–398.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator Leach, 1815 (Odonata: Aeshnidae) via NGS sequencing. Mitochondrial DNA B Resour. 1:783–786.
- Karjalainen S, Hämäläinen M. 2013. Demoiselle damselflies. winged jewels of silvery streams. Helsinki: Caloptera Publishing.
- Lee EM, Hong MY, Kim MI, Kim MJ, Park HC, Kim KY, Lee IH, Bae CH, Jin BR, Kim I. 2009. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: Ephemeridae) and Davidius lunatus (Odonata: Gomphidae). Genome. 52:810–817.
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa supports a basal Odonata within the Pterygota. Gene. 468:20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25:247–248.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41:353–358.
- Selva Kumar C, Nair RR, Sivaramakrishnan KG, Ganesh D, Janarthanan S, Arunachalam M, Sivaruban T. 2012. Influence of certain forces of evolution of synonymous codon usage bias in certain species of three basal orders of aquatic insects. Mitochondrial DNA. 23:447–460.
- Stewart JB, Beckenbach AT. 2006. Insect mitochondrial genomics 2: the complete mitochondrial genome sequence of a giant stonefly, Pteronarcys princeps, asymmetric directional mutation bias, and conserved plecopteran A + T-region elements. Genome. 49:815–824.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang JF, Chen MY, Chaw SM, Morii Y, Yoshimura M, Sota T, Lin CP. 2015. Complete mitochondrial genome of an enigmatic dragonfly, Epiophlebia superstes (Odonata, Epiophlebiidae). Mitochondrial DNA. 26:718–719.
- Wang L-J. 2000. Dragonflies of Taiwan. Taipei; JemJen Publishing.
- Yong HS, Song SL, Suana IW, Eamsobhana P, Lim PE. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Syst Ecol. 69:124–131.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2016. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2272–2273.
