Abstract
In this study, the complete mitochondrial genome of the Forcipomyia makanensis was sequenced and analyzed. The mitochondrial genome is 16,558 bp long and contains 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and one control region. Twenty-three genes were found to be encoded by the majority strand and the other 14 genes by minority strand, those is similar to that of other insects. The nucleotide compositing of the majority strands are 39.02% of A, 14.03% of C, 37.74% of T and 9.21% of G. The phylogenetic analysis by Maximum-likelihood (ML) method revealed that the Forcipomyia makanensis was close to the same family insect Culicoides arakawae.
Forcipomyia (Meigen (Diptera: Ceratopogonidae)), a genus with a worldwide distribution, is one of the species-richest in the biting midges, with many species being important pollinators of tropical and subtropical cultivated plants. Forcipomyia makanensis is one of the genus Forcipomyia, and the population number is very large in Makan village. However, little information about its genetic characteristic has been reported. Therefore, we determined to sequence the complete mitochondrial genome of F. makanensis using the De Novo sequencing techniques strategy, with the purpose to studying biogeographic, molecular, and population studies. The adult F. makanensis was obtained from bamboo forest in Makan, Zunyi, Guizhou, China (GPS 27.630765 N, 106.848949 E), and then were soaked in absolute ethanol. The specimens are deposited in Insect Collection of Zunyi Medical University, Guizhou Province, China (ICZU-CER-243). The adult F. makanensis were washing with 70% ethanol first, and then those were stored in 95% ethanol, the mitochondrial DNA was extracted using De Novo sequencing library, and DNA sequencing at BGI (Huada Gene Research Institute, Shenzhen, China).
The entire sequence of F. makanensis mitochondrial genome (Genbank accession no. MK000395) is 16558 bp in length, consisting of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA), and one control region. Twenty three genes (trnM, trnI, nad2,trnW, cox1, trnL2, cox2, trnK, trnD, atp8, atp6, cox3, trnG, nad3, trnA, trnR, trnN, trnS1, trnE, trnT, nad6, cytB, and trnS2) were found to be encoded by the majority strand and the other 14 genes (trnQ, trnC, trnY, trnF, nad5, trnH, nad4, nad4L, trnP, nad1, trnL1, l-rRNA, trnV, and s-rRNA) by minority strand, those is similar to that of other insects (Lin et al. Citation2017; Living Prairie Mitogenomics Consortium Citation2017; Singh et al. Citation2017). Overall nucleotide compositions of the majority strand are 39.02% of A, 14.03% of C, 37.74% of T and 9.21% of G, with an AT content of 76.76%.
With the except the cox1, all protein-coding genes with a typical ATN initiation codon, that is, ATG used in cox2, cox3, nad4, nad4L, cyt b and nad1, ATT in nad2, nad5 and nad6, ATA in atp6 and nad3, and ATC in atp8. Gene for cox1 used CAT as initiation codon. The cox3 showed an incomplete termination codon (TA), and the remaining 12 PCGs showed canonical stop codon pattern TAA. The lrRNA is located between tRNAL1 and tRNAV, whereas srRNA is accommodated between tRNAV and control region. The 22 tRNA genes vary from 61 to 72 bp in length. The secondary structure of tRNAs exhibited typical clover-leaf structure similar to other insect species. The size of the control region is 1549 bp (44.8%A, 3.36%G, 43.12% T, 8.72% C, with an AT content of 87.93%), and it located between s-rRNA and tRNAI, and the gene order around the control region is tRNAV-srRNA-control region-tRNAI-tRNAQ-tRNAM, and the order model was similar to other Diptera (Choudhary et al. Citation2015) and Coleopteran insects (Zhang and Hewitt Citation1997), and it is different to Lepidopteran insects (Singh et al. Citation2017).
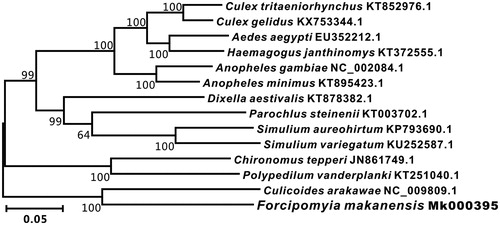
The phylogenetic tree of F. makanensis was constructed with the complete mtDNA sequences from 14 Diptera species by MEGA 7.0 (Kumar et al. Citation2016) using Maximum-likelihood (ML) methods (Liu et al. Citation2018). As shown in , the F. makanensis was close to Culicoides arakawae. Thus, this result supported the monophyly of F. makanensis.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Choudhary JS, Naaz N, Prabhakar CS, Rao MS, Das B. 2015. The mitochondrial genome of the peach fruit fly, Bactrocera zonata (Saunders) (Diptera: Tephritidae): complete DNA sequence, genome organization, and phylogenetic analysis with other tephritids using next generation DNA sequencing. Gene. 569:191–202.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lin ZQ, Song F, Li T, Wu YY, Wan X. 2017. New mitogenomes of two Chinese stag beetles (Coleoptera, Lucanidae) and their implications for systematics. J Insect Sci (Online). 17:63.
- Liu Q, Jiang X, Hou X, Cai R, Tan J, Chen W. 2018. The mitochondrial genome of Lasioderma serricorne (Coleoptera, Anobiidae). Mitochondrial DNA Part B. 3:64–65.
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA Part B. 2:344–346.
- Singh D, Kabiraj D, Sharma P, Chetia H, Mosahari PV, Neog K, Bora U. 2017. The mitochondrial genome of Muga silkworm (Antheraea assamensis) and its comparative analysis with other lepidopteran insects. PloS One. 12:e0188077.
- Zhang DX, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25:99–120.