Abstract
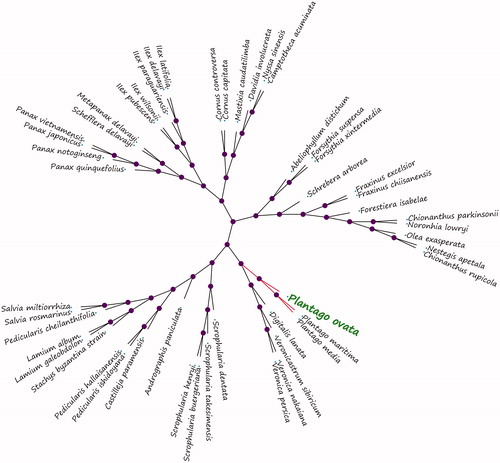
Plantago ovata is a commonly known and medicinally important plant of the family Plantaginaceae. It is more than 200 species of the family Plantago L. in the world. Simultaneously, Plantago ovata seed and husk are used in traditional medicine since long in China and treatment of cholesterol and diuresis. In this study, the complete chloroplast genome of Plantago ovata was determined by the Illumina Hiseq4000 sequencing. The complete chloroplast genome of Plantago ovata was 149,739 bp in length and displays a typical quadripartite structure of the large (LSC, 82,592 bp) and small (SSC, 18,365 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 24,390 bp each). It harbours 129 functional genes, including 86 protein-coding genes, 35 transfer RNA and 8 ribosomal RNA genes species. The overall nucleotide composition was: 30.4% A, 31.2% T, 19.5% C, and 18.9% G, with a total G + C content of 38.4%. Phylogenetic relationship analysis shows that Plantago ovata closely related to Plantago maritima and Plantago media.
Plantago ovata belongs to the Plantaginaceae and Plantago L. family. Plantago ovata is an economically and medicinally important medicinal herb in East Asia, particularly in China and India (Dhar et al. Citation2011). It is used extensively for the health and production of seed and husk for its application in pharmaceutical and food. In the pharmaceutical field, Plantago ovata has been used to treat cholesterol lowering, diuretic and others, it also has been incorporated into breakfast cereals, instant beverages, bakery and other dietary products in the food field (Kotwal et al. Citation2016). At present, there are few studies on the genome and transcriptome of P. ovata. In this study, we used the Illumina Hiseq4000 sequencing to get the complete chloroplast genome of Plantago ovata, in order to provide information for the study of the origin Medicinal plants of P. ovata in evolution.
The specimen of Plantago ovata was isolated from Jilin Agricultural University Jilin campus in Changchun, Jilin, China (125.41E; 43.81N) and the DNA of P. ovata was stored in Jilin Agricultural University College of Life Science (No. JLAUCLS6). The P. ovata DNA sample was sequenced as the paired end using the Hiseq4000 (Illumina Co., San Diego, CA). The P. ovata complete chloroplast genome was preliminarily annotated using the DOGMA (Dual Organellar GenoMe Annotator) online programme (Wyman et al. Citation2004), with default settings to identify protein-coding genes, rRNAs and tRNAs based on the Plant Plastid Code and BLAST homology searches. The secondary structures of transfer RNA (tRNA) genes were identified by using the ARAGORN (Laslett and Canback Citation2004) or through manually visual inspection.
The chloroplast genome sequence of P. ovata is a closed-circular molecule of 149,739 bp in length, consisting of 82,592 bp LSC region and 18,365 bp SSC region separated by a pair of 24,390 bp IR region. It displayed 129 functional genes set which observed in this plant chloroplast, including 86 PCGs, 35 tRNA genes (one for each amino acid, two each for Alanine, Asparagine, Glycine, Methionine and Threonine, three each for Isoleucine, Valine, Serine and Arginine, four each for Leucine), and 8 genes for ribosomal RNA subunits (two each for rrn16, rrn23, rrn4.5 and rrn5). The annotated chloroplast genome of P. ovata was submitted to GenBank of NCBI database under accession No. MH205737.
We selected other 49 related complete chloroplast genomes from GenBank to assess the phylogenetic relationship with P. ovata between them. The genome-wide alignment of all plant complete chloroplast genomes was done by HomBlocks (Bi et al. Citation2017). The phylogenetic trees were reconstructed using neighbor-joining (NJ) methods. NJ phylogenetic tree was constructed using MEGA-X with 5,000 bootstrap replicate (Kumar et al. Citation2018). All the nodes were inferred with strong support by the NJ methods. As shown in the phylogenetic tree (), Plantago ovata showed the closest with Plantago maritima and Plantago media.
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2017. HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics.110:18–22.
- Dhar MK, Kaul S, Sharma P, Gupta M. 2011. Plantago ovata: cultivation, genomics, chemistry and therapeutic applications. In: Singh RJ, editor. Genetic resources, chromosome engineering and crop improvement. New York, USA: CRC Press.
- Kotwal S, Kaul S, Sharma P, Gupta M, Shankar R, Jain M, Dhar MK. 2016. De novo transcriptome analysis of medicinally important Plantago ovata using RNA-Seq. Plos One. 11:e0150273.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.