Abstract
The grey-capped woodpecker (Dendrocopos canicapillus) is one species of family Picidae. In this study, the complete mitochondrial genome of D. canicapillus was determined and annotated for the first time. It is a circular molecule of 16,843 bp in size and contains 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and 2 control regions. The total base composition is 28.28% for A, 24.47% for T, 33.39% for C and 13.85% for G, respectively. A phylogenetic tree based on the DNA data of 13 PCGs using Neighbour Joining method was constructed to provide phylogenetic relationship between D. canicapillus and other species of Picidae.
Mitochondrial genome has become an important molecular marker for studies on population genetic and phylogenetic analyses because of its characteristics of maternal inheritance and fast mutation rate (Anmarkrud and Lifjeld Citation2017; Wada et al. Citation2010; Webb and Moore Citation2005). Grey-capped woodpecker (Dendrocopos canicapillus) is one species belonging to the genus Dendrocopos of family Picidae which includes more than 200 species and whose members are well known for their habit of pecking on wood to extract insects and their larvae (Fuchs et al. Citation2016; Shakya et al. Citation2017). Currently, only partial mitochondrial sequence for D. canicapillus is available. Here, we amplified and sequenced the complete mitochondrial DNA of D. canicapillus for the first time.
An adult D. canicapillus was collected from Jilin City of Jilin Province in China (44°00′31″N and 126°33′28″E) and stored in Zoological Specimen Museum of Jilin Agricultural University. Genomic DNA was extracted from muscle tissue using the standard phenol-chloroform protocol. The mitochondrial genome was amplified with 10 pairs of primers designed according to the known DNA sequences of D. canicapillus and Dendrocopos major. PCR products were purified and directly sequenced.
The complete mitochondrial genome DNA of D. canicapillus is a circular DNA molecule with a length of 16,843 bp containing 13 protein coding genes (PCGs), 2 ribosomal RNA (12S and 16S) genes, 22 transfer RNA genes (tRNAs) and 2 control regions (D-loop regions). The overall base composition is 28.28% for A, 33.39% for C, 13.85% for G and 24.47% for T, respectively. Except for ND6 and 8 tRNA genes, other genes are coded on the H strand. The mitochondrial genome of D. canicapillus was submitted to GenBank (accession no. MK015644).
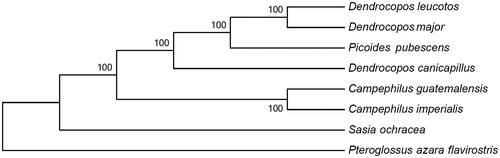
To explore the phylogenetic relation, the DNA data of 13 PCGs of D. canicapillus and other 7 Piciformes species were used to construct the phylogenetic tree using MEGA 5 with 1000 bootstrap replicates. As indicated by the neighbour-joining (NJ) tree (), the phylogenetic relationship between D. canicapillus and other Picidae species is consistent with that of the previous study (Eo Citation2017; Shakya et al. Citation2017). This complete mitogenome sequence of D. canicapillus will be useful for studying genetic diversity within this species and phylogenetic relationships among different Picidae species in the future.
Figure 1. The neighbour-joining (NJ) tree of 8 species was constructed based on the data set of 13 concatenated mitochondrial PCGs using MEGA 5 with 1000 bootstrap replicates. Sequence data used in this study are the following: Dendrocopos major (KT350609), Dendrocopos leucotos (KU131555), Picoides pubescens (KT119343), Campephilus imperialis (KU158198), Sasia ochracea (KT443919), Campephilus guatemalensis (KT443920), and Pteroglossus azara flavirostris (DQ780882).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Anmarkrud JA, Lifjeld JT. 2017. Complete mitochondrial genomes of eleven extinct or possibly extinct bird species. Mol Ecol Resour. 17:334–341.
- Eo SH. 2017. Complete mitochondrial genome of white-backed woodpecker Dendrocopos leucotos (Piciformes: Picidae) and its phylogenetic position. Mitochondrial DNA Part B. 2:451–452.
- Fuchs J, Pons J-M, Pasquet E, Bonillo C. 2016. Complete mitochondrial genomes of the white-browed piculet (Sasia ochracea, Picidae) and pale-billed woodpecker (Campephilus guatemalensis, Picidae). Mitochondrial DNA Part A. 27:3640–3641.
- Shakya SB, Fuchs J, Pons JM, Sheldon FH. 2017. Tapping the woodpecker tree for evolutionary insight. Mol Phylogenet Evol. 116:182–191.
- Wada K, Okumura K, Nishibori M, Kikkawa Y, Yokohama M. 2010. The complete mitochondrial genome of the domestic red deer (Cervus elaphus) of New Zealand and its phylogenic position within the family Cervidae. Anim Sci J. 81:551–557.
- Webb DM, Moore WS. 2005. A phylogenetic analysis of woodpeckers and their allies using 12S, Cyt b, and COI nucleotide sequences (class Aves; order Piciformes). Mol Phylogenet Evol. 36:233–248.
