Abstract
The complete mitochondrial genome of Setaria labiatepapillosa is reported for the first time in this study. Its entire mitogenome is 13,950 bp in length, contained 12 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and one noncoding regions. Arrangement of the 12 PCGs was identical to that of most filaria mtDNA sequence. The phylogenetic analyses by Bayesian inference method revealed that S. labiatepapillosa and S. digitata belong to Setariidae are in the same clade with strong support, the result supports classical taxonomic of the filariae. The mtDNA dataset provide a novel genetic marker for further studies of filaria.
Nematodes of the genus Setaria is prevalent world-wide which infects a variety of hosts such as cattle, sheep, equines, and pigs. The filaria are transmitted by various haematophagous arthropods (Singh et al. Citation2015). Setaria labiatepapillosa is one of the important members of genus Setaria, the adult parasites usually colonize the peritoneal surface and live free in the peritoneal cavity of hosts. When the larva stages infected may lead to cerebrospinal filariasis with neurological disorder symptom, which causing considerable economic losses to the live stock industries (Alasaad et al. Citation2012).
One adult filaria was collected from an infected sheep from Heilongjiang Province, China. The filaria was identified according to its morphological characteristics (Nakano et al. Citation2007) and stored in the Department of Parasitology, Heilongjiang institute of Veterinary Science (specimen no. CDL-170912). The total genomic DNA was extracted by TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), and stored at −20 °C until use. Primers were designed for PCR amplification and sequencing on the basis of the mitogenome sequence of Setaria digitata (GenBank accession No. GU138699) (Yatawara et al. Citation2010).
The entire S. labiatepapillosa mt genome was a typical circular DNA molecule with 13,950 bp in size (GenBank accession number MH937750). There were twelve PCGs (cox1-3, nad1-6, nad4L, atp6, and cytb), twenty-two transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one noncoding regions (NCR) including in the mtDNA sequence of S. labiatepapillosa. Arrangement of the 12 PCGs was identical to that of most filaria mtDNA sequence (Hu et al. Citation2003; Yatawara et al. Citation2010). The nucleotide contents of the mt genome sequence were biased toward A + T (79.0%), in accordance with the filaria mt genomes sequenced to date (Ramesh et al. Citation2012). The S. labiatepapillosa mt genome encoded 3461 amino acids in total. The A + T contents of PCGs ranged from 72.1% (cox1) to 87.2% (nad6). Codon usage analyses of S. labiatepapillosa showed that four kinds of initiation codons (ATT, ATA, ATG and TTG) and three kinds of termination codons (TAG, TAA, and T) were used. The size of the 22 tRNAs ranging from 53 to 69 bp. The rrnL (951 bp) is located between trnH and nad3, while rrnS (679 bp) is located between nad4L and trnY. The NCR (495 bp) is located between cox3 and trnA, which A + T contents are 92.7%.
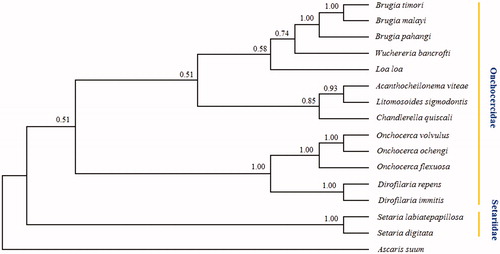
Phylogenetic analyses of Filarioidea using bayesian inference (BI) methods based on the concatenated amino acid sequences of 12 PCGs with ascaris suum as outgroup. Phylogenetic analyses revealed that Onchocercidae and Setariidae formed two clades. Within Onchocercidae, genera Onchocerca grouped with genera Dirofilaria in the same branch, and others filaria (genera Brugia, Wuchereria, Loa, Acanthocheilonema, Litomosoides, and Chandlerella) in the other branch. Within Setariidae, S. digitata and S. labiatepapillosa are in the same clade with strong support, this result supports classical taxonomic of the filariae (). The mtDNA dataset should provide a novel genetic marker for further studies of the taxonomy, identification and population genetics of these and other related filaria of socio-economic significance.
Figure 1. Phylogenetic relationships of Setaria labiatepapillosa and other 14 species filaria based on concatenated amino acid sequences of 12 protein-coding genes were analyzed with Bayesian inference (BI) method, using Ascaris suun (NC_001327) as an outgroup. GenBank accessions for sequences are: Acanthocheilonema viteae, AP017679; Brugia malayi, AF538716; Brugia pahangi, AP017680; Brugia timori, AP017686; Chandlerella quiscali, HM773029; Dirofilaria immitis, AJ537512; Dirofilaria repens, KR071802; Litomosoides sigmodontis, AP017689; Loa loa, HQ186250; Onchocerca flexuosa, HQ214004; Onchocerca ochengi, AP017693; Onchocerca volvulus, AP017695; Setaria digitata, GU138699; Wuchereria bancrofti, JN367461; Setaria labiatepapillosa in present study, MH937750.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Alasaad S, Pascucci I, Jowers MJ, Soriguer RC, Zhu XQ, Rossi L. 2012. Phylogenetic study of Setaria cervi based on mitochondrial cox1 gene sequences. Parasitol Res. 110:281–285.
- Hu M, Gasser RB, Abs EI, Osta YG, Chilton NB. 2003. Structure and organization of the mitochondrial genome of the canine heartworm, Dirofilaria immitis. Parasitology. 127:37–51.
- Nakano H, Tozuka M, Ikadai H, Ishida H, Goto R, Kudo N, Katayama Y, Muranaka M, Anzai T, Oyamada T. 2007. Morphological survey of bovine Setaria in the abdominal cavities of cattle in Aomori and Kumamoto Prefectures, Japan. J Vet Med Sci. 69:413–415.
- Ramesh A, Small ST, Kloos ZA, Kazura JW, Nutman TB, Serre D, Zimmerman PA. 2012. The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Mol Biochem Parasitol. 183:32–41.
- Singh H, Singh NK, Singh ND, Jyoti Rath SS. 2015. Occurrence of Setaria labiatopapillosa in peritoneal cavity of a crossbred cattle. J Parasit Dis. 39:152–154.
- Yatawara L, Wickramasinghe S, Rajapakse RP, Agatsuma T. 2010. The complete mitochondrial genome of Setaria digitata (Nematoda: Filarioidea): mitochondrial gene content, arrangement and composition compared with other nematodes. Mol Biochem Parasitol. 173:32–38.
