Abstract
Homalium (Salicaceae) is mainly distributed in tropical and subtropical regions. Here we report and characterize the complete plastid genome sequences of Homalium paniculiflorum, H. stenophyllum, and H. ceylanicum in an effort to provide genomic resources useful for promoting its systematics research. The plastomes of H. paniculiflorum, H. stenophyllum, and H. ceylanicum were found to possess a total length 157,206 bp, 156,395 bp, and 156,551 bp, respectively. They all have typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 27,796 bp in H. paniculiflorum, 27,669 in H. stenophyllum and 27,639 bp in H. ceylanicum, a Large Single-Copy (LSC) region of 85,031 bp (H. paniculiflorum), 84,458 bp (H. stenophyllum) and 84,627 bp (H. ceylanicum), a Small Single-Copy (SSC) region of 16,584 bp (H. paniculiflorum), 16,599 bp (H. stenophyllum), and 16,646 bp (H. ceylanicum). The all plastome contains 111 genes, consisting of consisting of 77 unique protein-coding genes (seven of which are duplicated in the IR), 30 unique tRNA genes (seven of which are duplicated in the IR) and 4 unique rRNA genes. The overall A/T content in the plastomes of H. paniculiflorum, H. stenophyllum, and H. ceylanicum are 63.30, 63.20, and 63.20%. The complete plastome sequences of H. paniculiflorum, H. stenophyllum, and H. ceylanicum will provide a useful resource for the conservation genetics of the three species as well as for the phylogenetic studies of Homalium.
Homalium (Salicaceae) is mainly distributed in tropical and subtropical regions. Most of Homalium species could be used for furniture. In this study, H. paniculiflorum and H. stenophyllum were sampled from Diaoluo Mountain (18.67°N, 109.88°E) National Nature Reserve of Hainan, China. H. ceylanicum was sampled from Jianfeng Mountain (18.74°N, 108.86°E) National Nature Reserve of Hainan, China. The voucher specimens (H. paniculiflorum, specimen code: Wang et al., B13 thereafter; H. stenophyllum, Wang et al., B17 and H. ceylanicum, Wang et al., B88) were deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Populus lasiocarpa (NC_036040.1) (Zhang Citation2017) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters, Auckland, New Zealand) against the plastome of Populus lasiocarpa (NC_036040.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastomes of H. paniculiflorum, H. stenophyllum, and H. ceylanicum were found to possess a total length 157,206 bp, 156,395 bp, and 156,551 bp, respectively. They all have typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 27,796 bp in H. paniculiflorum, 27,669 in H. stenophyllum and 27,639 bp in H. ceylanicum, a Large Single-Copy (LSC) region of 85,031 bp (H. paniculiflorum), 84,458 bp (H. stenophyllum) and 84,627 bp (H. ceylanicum), a Small Single-Copy (SSC) region of 16,584 bp (H. paniculiflorum), 16,599 bp (H. stenophyllum) and 16,646 bp (H. ceylanicum). The all plastome contains 111 genes, consisting of consisting of 77 unique protein-coding genes (seven of which are duplicated in the IR), 30 unique tRNA genes (seven of which are duplicated in the IR) and 4 unique rRNA genes. The overall A/T content in the plastomes of H. paniculiflorum, H. stenophyllum and H. ceylanicum are 63.30, 63.20, and 63.20%. The first species’s corresponding value of the LSC, SSC, and IR region were 65.40, 69.50, and 58.10%, the second one were 65.30, 69.40, and 58.10%, respectively, the last one were 65.40, 69.40, and 58.00%, respectively.
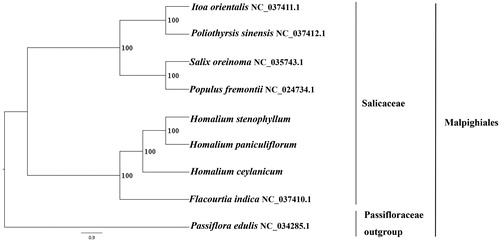
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of five published complete plastomes of Salicaceae, using Passiflora edulis (Passifloraceae) as an outgroup. The phylogenetic analysis indicated that H. paniculiflorum, H. stenophyllum, and H. ceylanicum are close to Flacourtia indica within Salicaceae (). Most nodes in the plastome ML tree were strongly supported. The complete plastome sequences of H. paniculiflorum, H. stenophyllum, and H. ceylanicum will provide useful resources for the conservation genetics of the three species as well as for the phylogenetic studies of Homalium.
Figure 1. The best ML phylogeny recovered from 9 complete plastome sequences by RAxML. Accession numbers: Homalium paniculiflorum (this study, GenBank accession number: MK033523), Homalium stenophyllum (this study, GenBank Accession number: MK033524), Homalium ceylanicum (this study, GenBank accession number: MK033525), Itoa orientalis NC_037411.1, Poliothyrsis sinensis NC_037412.1, Salix oreinoma NC_035743.1. Populus fremontii NC_024734.1, Flacourtia indica NC_037410.1; outgroup: Passiflora edulis NC_034285.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models . Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang L. 2017. Complete chloroplast genome of Populus lasiocarpa. Submitted (18-OCT-2017) Bethesda, MD (USA): National Center for Biotechnology Information, NIH.
