Abstract
Rosemary (Rosmarinus officinalis) is a perennial herb with fragrant endemic to Mediterranean region. Despite its important economic values on flavour, food and medicinal industry, the whole chloroplast (cp) genome of it has not been reported yet. Here, the complete chloroplast genome of the R.officinalis has been reconstructed from the whole-genome Illumina sequencing data. The circular genome is 152,462 bp in size, and comprises a pair of inverted repeat (IR) regions of 25,569 bp each, a large single-copy (LSC) region of 83,355 bp and a small single-copy (SSC) region of 17,969 bp. The overall GC content of the cp genome was 38.0%. The chloroplast genome contains 132 genes, including 94 protein-coding genes, 30 transfer RNA genes, and eight ribosomal RNA genes. Phylogenomic analysis strongly supported the close relationship of R. officinalis and Scutellaria baicalensis.
Rosemary (Rosmarinus officinalis) is a perennial herb, which is woody, evergreen with needle-like leaves and white, purple or blue flowers. It is native to the Mediterranean and Asia. Rosemary essential oils was used in bodily perfumes industry, and also used as a flavouring in foods. In addition, rosemary is reported to have antimicrobial and antioxidant activity and could be developed health food, drugs or natural insecticides (Jiang et al. Citation2011; Ibañez et al. Citation2003). Therefore, it is highly necessary to survey the genetic background. In this study, we assembled and characterized the complete chloroplast genome sequence of R. officinalis using the Illumina sequencing reads.
DNA samples were extracted from the fresh leaves that were collected from a single individual of R. officinalis in Xi’an Botanical Garden of Shaanxi Province (34°12.613′N, 108°57.091′E). The whole genome shotgun sequencing of R. officinalis was performed by Beijing Gene Insititution (BGI, Shenzhen, China) using the Illumina HiSeq 2500 platform (Illumina, Hayward, CA). Total 36.1 M 125 bp raw reads were retrieved and trimmed by CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark). A subset of 16.72 M trimmed reads were used for reconstructing the chloroplast genome by MITObim v1.8 (Hahn et al. Citation2013), with that of its congener Sesamum indicum (GenBank: NC_016433.1) as the initial reference genome. A total of 13,543,658 individual chloroplast reads yielded an average coverage of 526-fold. The chloroplast genome was annotated in GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand) and was drawn to the circular chloroplast genome sequence map of OGDRAW.
The chloroplast genome of R. officinalis is a double-stranded circular DNA molecule with 152,462 bp in size (NC_027259). It comprises a pair of inverted repeat (IR) regions of 25,569 bp each, separated by a large single-copy (LSC) region of 83,355 bp and a small single-copy (SSC) region of 17,969 bp. The total GC content is 38%, however the GC content is unevenly distributed in the entire cp genome. It is highest in the IR regions (43.0%), median in the LSC region (36.2%) and lowest in the SSC regions (31.9%). This is similar to those previously reported for the chloroplast genomes of most other vascular plants (Yang et al. Citation2014).
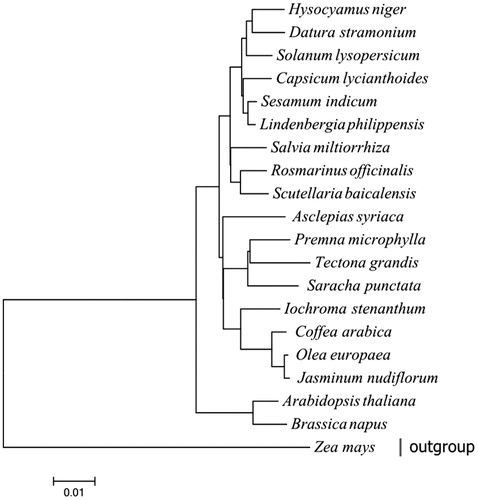
This chloroplast genome harbours 132 functional genes, including 94 protein-coding genes (PCGs), 30 tRNA genes and eight rRNA genes. 41 PCGs, 17 tRNA genes and 4 rRNA genes are located in the forward strand while others are located in the reverse strand. Moreover, 9 PCGs, 6 tRNA, and all rRNA are duplicated in the IR regions. The LSC region contains 64 PCGs and 17 tRNA genes but one tRNA and 12 PCGs in the SSC region. Among them, 39 genes are involved in photosynthesis and 26 genes are related with self-replication. As shown in the Maximum-Likelihood phylogenetic tree (), R. officinalis is sister to Scutellaria baicalensis, and this close relationship received a strong bootstrap support. Our findings will provide a foundation for further investigation of chloroplast genome evolution in Lamiaceae.
Figure 1. The Maximum-likelihood phylogenetic tree was generated using 39 shared PCGs among 20 chloroplast sequences by MEGA 6.0 using 500 bootstrap replicates. The analyzed species and corresponding Genbank accession numbers are as follows: Arabidopsis thalina (NC_000932), Asclepias syriaca (NC_022432), Brassica napus (NC_016734), Capsicum lycianthoides (KP274856), Coddea arabica (NC_008535), Datura stramonium (NC_018117), Hyoscyamus niger (NC_024261), Iochroma loxense (KP296185), Jasminum nudiflorum (NC_008407), Lindenbergia philippensis (NC_022859), Olea europaea (NC_013707), Premna microphylla (NC_026291), Rosmarinus officinalis (NC_027259), Saracha punctata (KP280050), Scutellaria baicalensis (NC_027262), Sesamum indicum (NC_016433), Solanum lysopersicum (NC_007898), Tectona grandis (NC_020098), Zea mays (NC_001666).
Disclosure statement
The authors report no conflicts of interest, and alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Ibañez E, Kubátová A, Señoráns FJ, Cavero S, Reglero G, Hawthorne SB. 2003. Subcritical water extraction of antioxidant compounds from rosemary plants. J Agric Food Chem. 51:375–382.
- Jiang Y, Wu N, Fu YJ, Wang W, Luo M, Zhao CJ, Zu YG, Liu XL. 2011. Chemical composition and antimicrobial activity of the essential oil of Rosemary. Environ Toxicol Pharmacol. 32:63–68.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
