Abstract
The mitochondrial genome of the seed pest from Sophora moorcroftiana was completely determined. The mitochondrial genome was 15,294 bp in length and contained 37 typical invertebrate mitochondrial genes, including 13 protein-coding genes (PCG), two rRNA genes, 22 tRNA genes and one control region. All polyproteins started with Met. All the PCGs harbored the termination codon TAA except the incomplete termination codon T for COII gene. All tRNAs can folded into standard cloverleaf secondary structures. Phylogenetic analysis showed that the seed pest from Sophora moorcroftiana is more closely related to a newly reported species in Meroptera genus (under GenBank no. MF073207).
Sophora moorcroftiana, a perennial endemic leguminous low shrub in Tibet, which plays an important role in vegetation restoration, is found in valleys, slopes, terraces from 3000 to 3900 m above sea level along the middle reaches of the Yarlung Zangbo River (Liu et al. Citation2006; Zhao et al. Citation2007). However, the seed of Sophora moorcroftiana is facing severe pest challenge. A specie from snout moth (Insecta: Lepidoptera: Pyralidae) was considered to be the major destructive pest and its larva can damage seed by increasing moisture content and contaminating it with faces and dead bodies (Wang et al. Citation2010). The larva of this specie is morphologically highly similar with other snout moth, while the adults are tiny and hardly investigated using routine field surveys and observations (Hrcek et al. Citation2011). No reports about genome or mitogenome of this species. Hence, we selected the mitogenomic sequences as a tool for studying it for the ease of recovering genetic information (Jiang et al. Citation2005; Bossuyt et al. Citation2006).
Samples of snout moth associated with Sophora moorcroftiana seeds (SMASM) were collected in the semi-arid valley of Lhasa in the Tibetan Plateau, China, and deposited in the Insect Collection of XiZang Agricultural and Animal Husbandry College. Larva was used for DNA extraction, and then PCR amplification, sequencing and assembling were conducted. Finally, we determined the complete mitogenome of SMASM was 15,294 bp in length (GenBank accession no. MG748599). It contained 37 typical mitochondrial genes including 13 proteins coding genes (PCGs), 22 tRNAs genes, two rRNAs together with a control region (CR) (Boore et al. Citation1998; Kim et al. Citation2006).
Eleven proteins genes were initiated with typical ATN (five with ATG, five with ATT, one with ATA). The start codon of COI gene was CGA, which was the same as other species (Liao et al. Citation2010) and a unique initiator of AAA was detected for ND4L gene. However, all the proteins started with Met, and the 13 PCGs harbored the termination codon TAA except the incomplete termination codon T for COII gene. The Relative Synonymous Codon Usage (RSCU) in PCGs was analyzed and results indicated one absent codon of CGG and five most frequently used codons, AUU (I), AAU (N), UUU (F), UUA (A) and AUA (M), in SMASM mitogenome. All tRNAs can folded into standard cloverleaf secondary structures.
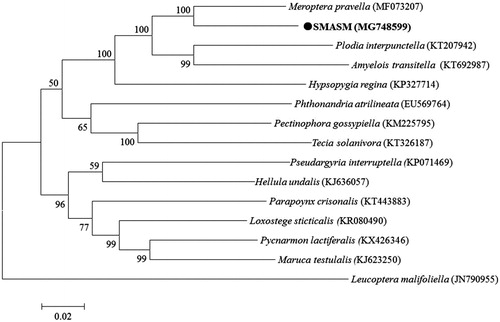
Phylogenetic analysis () showed with strong bootstrap probability of (100%) that SMASM had a closer relationship with Meroptera pravella (Jeffrey Citation2017). Homologous analysis also indicated that SMASM had a highest identity of 91.1% with Meroptera pravella. The results suggested that SMASM should be a species of Meroptera genus, family Pryradidae. This is the first report of species from Meroptera genus in China. The newly determined mitogenome of SMASM will help to understand the evolution of insects in Pryradidae.
Figure 1. A maximum-likelihood tree illustrating the phylogenetic position of SAMAS among other species in family Pryradidae. The maximum-likelihood analysis was conducted using the whole mitogenomic sequences. Numbers of each node are bootstrap probabilities by 500 replications shown only when they are 50% or larger. The GenBank accession numbers of the mitogenomic sequences for each taxon are in parentheses.
Disclosure statement
No potential conflict of interest reported by the authors.
Additional information
Funding
References
- Boore JL, Lavrov D, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 393:667–668.
- Bossuyt F, Brown RM, Hillis DM, Cannatella DC, Milinkovitch MC. 2006. Phylogeny and biogeography of a cosmopolitan frog radiation: late cretaceous diversification resulted in continent-scale endemism in the family ranidae. Syst Biol. 55:579.
- Hrcek JAN, Miller SE, Quicke DL, Smith M. 2011. Molecular detection of trophic links in a complex insect host-parasitoid food web. Mol Ecol Resour. 11:786–794.
- Jeffrey MM. 2017. The complete mitochondrial genome of the lesser aspen webworm moth meroptera pravella (insecta: lepidoptera: pyralidae). Mitochondrial DNA Part B: Resour. 2:344–346.
- Jiang J, Dubois A, Ohler A, Tillier A, Chen X, Xie F, Stöck M. 2005. Phylogenetic relationships of the tribe paini (amphibia, anura, ranidae) based on partial sequences of mitochondrial 12s and 16s rRNA genes. Zool Sci. 22:353–362.
- Kim I, Lee EM, Seol KY, Yun EY, Lee YB, Hwang JS, Jin BR. 2006. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol. 15:217–225.
- Liao F, Wang L, Wu S, Li YP, Zhao L, Huang GM, Niu CJ, Liu YC, Li MG. 2010. The complete mitochondrial genome of the fall webworm, hyphantria cunea (lepidoptera: arctiidae). Int J Biol Sci. 6:172–186.
- Liu ZM, Zhao AM, Kang XY, Zhou SL, López-Pujol J. 2006. Genetic diversity, population structure, and conservation of Sophora moorcroftiana (Fabaceae), a shrub endemic to the Tibetan Plateau. Plant Biol (Stuttg). 8:81–92.
- Wang WJ, Fang JP, Liu YC, Wang B. 2010. The spatial distribution model of etiella zinckenella larvae in Sophora moorcroftiana of Tibet. J Henan Agric Sci. 39:83–87.
- Zhao W, Zhang Z, Li Q. 2007. Growth and reproduction of Sophora moorcroftiana responding to altitude and sand burial in the middle Tibet. Environ Geol. 53:11–17.
