Abstract
In the present study, we determined the complete mitogenome sequence of scleractinia, Goniopora djiboutiensis using Illumina HiSeq platform. The assembled mitogenome was 18,765 bp in length, comprising unique 13 protein-coding genes (PCGs), two ribosomal RNAs, and two transfer RNAs genes of each, showing a typical scleractinia pattern. New phylogenetic analysis upon complete mitogenomics revealed that G. djiboutiensis is most closely related to G. columna, with high bootstrap values. It would provide further insights on the taxonomy and phylogeny of genus Goniopora de Blainville.
Increasing atmospheric carbon dioxide concentration contributes to global warming and alters ocean carbonate chemistry in the process known as ocean acidification. It increases the potential for dissolution of present skeletons (Edmunds and Yarid Citation2017; Kenkel et al. Citation2017). Variable skeletal morphology, genotype-induced plasticity, and homoplasy of skeletal structures have presented major challenges for scleractinian coral taxonomy and systematics (Terraneo et al. Citation2016). Mitochondria are highly conserved and differ from nuclear genes. The rate of evolution makes it an ideal tool for studying evolution and molecular ecology (Jiang et al. Citation2017; Wang et al. Citation2018).
The genus Goniopora de Blainville, 1830, common across the Indo-Pacific, is characterized by a genus-specific septal formula. Species identification to date has been conducted most using traditional macromorphology, such as colony growth form, corallite dimensions, and number and fusion pattern of septa (Terraneo et al. Citation2016; Gonzálezespinosa et al. Citation2018; Edinger and Risk Citation2000). However, it is taxonomic uncertainty due to confusing patterns of morphological variation, with surprising examples of convergent evolution, rapid evolution and phenotypic plasticity (Tisthammer and Richmond Citation2018).
In present study, the complete mitochondrial genome of Goniopora djiboutiensis was newly determined. After morphological identification, the specimens were snap-frozen stored in liquid nitrogen within 10 min of collection until further processing.
Samples (voucher no. DA01) of G. djiboutiensis were collected from Daao Bay in Guangdong, China (114°28′02.10″E, 22°33′06.21″N) on November 2017. We used the high-throughput sequencing method to acquire the G. djiboutiensis complete mitochondrial genome sequences. The raw next generation sequencing reads generated from HiSeq X-ten (Illumina, San Diego, CA). The reads were de novo assembly by using commercial software (SPAdes V3.9.0, St Petersburg, Russian) to produce a single, circular form of complete mitogenome with about an average 237.9 coverage. The complete mitogenome of G. djiboutiensis was 18,765 bp in size (GenBank MH746816) and its overall base composition is 25.6% for A, 13.7% for C, 23.5% for G, and 37.2% for T, and have GC content of 37.2%, showing 99% identities to Goniopora columna (GenBank JF825141.1). The protein coding, rRNA, and tRNA genes of G. djiboutiensis mitogenome were predicted by using blastn, UGENE V1.30 (UGENE, Novosibirsk, Russia), RNAmmer 1.2 Server tools, and manually inspected. The complete mitogenome of G. djiboutiensis includes unique 13 protein-coding genes (PCGs), two transfer RNA genes (tRNAMet, tRNA-Trp), and two ribosomal RNA genes. Among the 13 PCGs, the longest one is ND5 gene (1836 bp), whereas the shortest is ATP8 gene (216 bp). The ND5 gene is split into two parts by a large fragment of genes, which commonly presented in scleractinian coral, and the size of inserted fragment was usually over 10 Kb.
All PCGs, tRNA, and rRNA genes were encoded on H-strand. The most used start codons is ATG. There are 8 PCGs started with ATG codon (ND1, Cyt b, ND2, ATP6, COX3, COX2, COX1, and ATP8), three GTG codon (ND5, ND4L and ND3), one with ATT codon (ND6), and one with ATA codon (ND4). Seven of the 13 PCGs are inferred to terminate with TAA (ND4, ND1, ND2, ND6, ATP8, ND4L, and COX1), six with TAG (ATP6, Cytb, ND3, ND5, COX2 and COX3). The small and large mitochondrial ribosomal RNA genes of G. djiboutiensis were located opposite each other on the circular genome as in other corals (Ju et al. Citation2017).
To determine the phylogenetic position of G. djiboutiensis, we used MEGA Version10.0.4 (Gene Codes, Ann Arbor, MI) software to construct a Maximum likelihood tree using 500 bootstrap replicates and Kimura 2-parameter model (Tian and Niu Citation2017).The phylogenetic tree was reconstructed with 11 coral species complete mitogenomes derived from GenBank. Paracorallium japonicum was used as an outgroup for tree rooting ().
Figure 1. Molecular phylogeny of Goniopora djiboutiens and related species in Scleractinia based on complete mitogenome. The complete mitogenomes are downloaded from GenBank and the phylogenetic tree is constructed by maximum-likelihood method with 500 bootstrap replicates. The gene's accession number for tree construction is listed behind the species name.
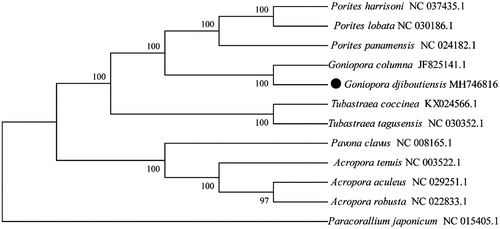
The result showed G. djiboutiens was grouped into a single clade with G. columna. This relationship has been verified by molecular phylogenies, which were with 100% bootstrap value supported. In conclusion, the complete mitogenome of G. djiboutiensis determined the essential phylogenetic and evolutionary information of corals in this study.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Edinger EN, Risk MJ. 2000. Reef classification by coral morphology predicts coral reef conservation value. Biol Conserv. 92:1–13.
- Edmunds PJ, Yarid A. 2017. The effects of ocean acidification on wound repair in the coral Porites spp. J Exp Marine Biol Ecol. 486:98–104.
- Gonzálezespinosa PC, Pazgarcía DA, Reyesbonilla H, Cabraltena RA, Balart EF. 2018. Evidence of sexual dimorphism in skeletal morphology of a gonochoric reef coral. Royal Soc Open Sci. 5:1–7.
- Jiang L, Li Z, Cheng D, Zhu L, Min Z, Ruan Q, Wei C. 2017. The complete mitochondrial genome sequence of the Sichuan Digging Frog, Kaloula rugifera (Anura: Microhylidae) and its phylogenetic implications. Gene. 626:367–375.
- Ju YM, Hsiao ST, Kuo FW, Wu JH. 2017. The complete mitochondrial genome of Montipora aequituberculata (Scleractinia, Acroporidae). Mitochondrial DNA Part B. 2:62–63.
- Kenkel CD, Moya A, Strahl J, Humphrey C, Bay LK. 2017. Functional genomic analysis of corals from natural CO2 – seeps reveals core molecular responses involved in acclimatization to ocean acidification. Global Change Biol. 24:1–14.
- Terraneo TI, Benzoni F, Arrigoni R, Berumen ML. 2016. Species delimitation in the coral genus Goniopora (Scleractinia, Poritidae) from the Saudi Arabian Red Sea. Mol Phylogenet Evol. 102:278–294.
- Tian P, Niu W. 2017. The complete mitochondrial genome of the Acropora pruinosa. Mitochondrial DNA Part B. 2:652–653.
- Tisthammer KH, Richmond RH. 2018. Corallite skeletal morphological variation in Hawaiian Porites lobata. Coral Reefs. 37:1–12.
- Wang X, Tian P, Niu W, Yu S. 2018. The complete mitochondrial genome of the Montipora peltiformi (Scleractinia: Acroporidae). Mitochondrial DNA Part B. 3:99–100.
