Abstract
The complete chloroplast genome sequences of newly discovered cliff-dwelling species of Sonchus, S. boulosii, were reported in this study. The S. boulosii plastome was 152,016 bp long, with the large single copy (LSC) region of 83,988 bp, the small single copy (SSC) region of 18,566 bp, and two inverted repeat (IR) regions of 24,731 bp. The plastome contained 130 genes, including 88 protein-coding, six ribosomal RNA, and 36 transfer RNA genes. The overall GC content was 31.2%. Phylogenetic analysis of 12 representative plastomes within the order Cichorieae suggests that S. boulosii is closely related to S. oleraceus.
The genus Sonchus L. belongs to the subtribe Hyoseridinae (formerly Sonchinae) of Cichorieae and, based on a new and broad concept of the genus, it includes approximately 90 species in eight distinct clades (Kim et al. Citation2007; Kilian et al. Citation2009). Sonchus s.l. includes not only some of the most spectacular examples of adaptive radiation on oceanic islands in the Atlantic (the woody Sonchus alliance in the Macaronesian Islands) and the Pacific (Dendroseris in the Juan Fernández Islands) but also some widely distributed weedy species (e.g. S. asper, S. oleraceus, S. arvensis, etc.) (Kim et al. Citation1996). Recently, a distinct new lineage of Sonchus was discovered in Morocco and, based on morphological and molecular divergence from other sections, it was subsequently recognized as a new section Pulvinati (Mejías et al. Citation2018). Sonchus boulosii, as a sole representative of new section Pulvinati, has been found only in four populations from three distinct biogeographic regions of Morocco (i.e., the Eastern Morocco mountains, the Middle Atlas, and the High Atlas). It is a very rare cliff-dwelling species and hypothesized to be quite old relic member within Sonchus, with sharing some primitive characteristics with section Pustulati. Although phylogenetic relationships among major lineages within Sonchus s.l. and closely related genera (Reichardia and Launaea) have been investigated rather thoroughly, additional resolutions and robust supports within and among lineages are warranted (Kim et al. Citation1996, Citation2007; Mejías et al. Citation2018). Furthermore, very little is known for chloroplast genome evolution within the genus Sonchus and the subtribe Hyoseridinae (Hereward et al. Citation2018). As an initial step to investigate the chloroplast genome evolution within this group, we reported the complete chloroplast genome of S. boulosii and assessed phylogenetic position within Cichorieae.
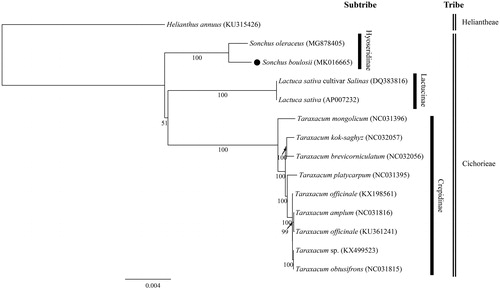
Figure 1. The maximum-likelihood (ML) tree based on the 12 representative chloroplast genomes of Cichorieae. The bootstrap value based on 1000 replicates is shown on each node.
Total DNA (Voucher specimen: 32°35′11.5″N 4°08′55.7″W, ECWP s.n.) was isolated using the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and sequenced by the Illumina HiSeq 4000 (Illumina Inc., San Diego, CA). A total of 51,623,340 paired-end reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). RNAs (rRNA) were identified using RNAmmer 1.2 Server (Lagesen et al. Citation2007) and the transfer RNAs (tRNA) were predicted using ARAGORN v 1.2.36 (Laslett and Canback Citation2004).
The total plastome length of S. boulosii (GenBank accession number: MK016665) was 152,016 bp, with large single copy (LSC; 83,988 bp), small single copy (SSC; 18,566 bp), and two inverted repeats (IRa and IRb; 24,731 bp each). The overall GC content was 37.6% (LSC, 35.8%; SSC, 31.2%; IRs, 43.1%) and the plastome contained 130 genes, including 88 protein-coding, six rRNA, and 36 tRNA genes. Two major inversions reported in Asteraceae were also found in S. boulosii (Timme et al. Citation2007). To confirm the phylogenetic position of S. boulosii, 12 representative species of Cichorieae were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015). The ML tree () showed that S. boulosii is sister to S. oleraceus.
Acknowledgments
The initial discovery phase of this study was funded by the Emirates Center for Wildlife Propagation (ECWP), a project of the International Fund for Houbara Conservation (IFHC). Thus, we are grateful to H. H. Sheikh Mohammed bin Zayed Al Nahyan, Crown Prince of Abu Dhabi and Chairman of the IFHC, and S. E. Mohammed Al Bowardi, Deputy Chairman of IFHC, for their support. It was also conducted under the guidance of RENECO International Wildlife Consultants, LLC, a consulting company managing ECWP. We are thankful to Frédéric Lacroix, managing director, and Gwénaëlle Levèque, project director, for their supervision.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Disclosure Statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Hereward JP, Werth JA, Thornby DF, Keenan M, Chauhan BS, Walter GH. 2018. Complete chloroplast genome of glyphosate resistant Sonchus oleraceus L. from Australia, with notes on the small single copy (SSC) region orientation. Mitochondrial DNA B. 3:363–364.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kilian N, Gemeinholzer B, Lack HW. 2009. 24. Cichorieae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, editors. Systematics, evolution and biogeography of Compositae. Austria: International Association for Plant Taxonomy, University of Vienna; pp. 343–383.
- Kim S-C, Crawford DJ, Jansen RK. 1996. Phylogenetic relationships among the genera of the subtribe Sonchinae (Asteraceae): evidence from ITS sequences. Syst Bot. 21:417–432.
- Kim S-C, Lee C, Mejías JA. 2007. Phylogenetic analysis of chloroplast DNA matK gene and ITS of nrDNA sequences reveals polyphyly of the genus Sonchus and new relationships among the subtribe Sonchinae (Asteraceae: Cichorieae). Mol Phylogenet Evol. 44:578–597.
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNammer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108.
- Laslett D, Canback B. 2004. ARAGORN, a program for the detection of transfer RNA and transfer-messenger RNA genes. Nucleic Acids Res. 32:11–16.
- Mejías JA, Chambouleyron M, Kim S-H, Dolores Infante M, Kim S-C, Léger J-F. 2018. Phylogenetic and morphological analysis of a new cliff-dwelling species reveals a remnant ancestral diversity and evolutionary parallelism in Sonchus (Asteraceae). Plant Syst Evol. 304:1023–1040.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Timme RE, Kuehl JV, Boore JL, Jansen RK. 2007. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 94:302–312.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
