Abstract
We sequenced almost complete mitochondrial genome of Apis dorsata (Insecta: Hymenoptera: Apocrita: Apidae) with a length of 15,933 bp. The mitogenome sequenced harbours 13 protein-coding genes (PCGs), 21 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA), and one control region (D-loop) with very high AT-base content. The genome has similar codon usage and gene organization to those of mitogenome reported for other Hymenoptera. Phylogenetic analyses showed that Apis dorsata and (Apis cerana+Apis mellifera) are the sister taxa, and more original, which is widely accepted view.
Apis dorsata are the largest Asian honeybees in size and have the most matings per queen, occurs from western India, eastwards below the Himalayas, throughout continental Asia and oceanic Asia, including the Philippines and Sulawesi, Indonesia. In terms of altitudinal distribution, most are below 1000 m and the other between 1000 m and 3000 m (Hepburn et al. Citation2005). In this study, we presented the mitochondrial genome of the Apis species, Apis dorsata, which belong to Apinae. Although A. dorsata and A. laboriosa are closely, significant difference exists in the altitudinal distributions between A. dorsata and A. laboriosa, and, while they are partially sympatric, appear to be seasonally isolated (Otis Citation1996; Thapa et al. Citation2001).
The tissue sample of A. dorsata was collected from JiNuo town, Jing hong City, Yunnan province, southwest China (coordinate as follows: N22°04.602′E101°00.920), and the specimens were deposited at Eastern Bee Research Institute, Yunnan Agricultural University, in Kunming, Yunnan, China. Genetic DNA from head and chest of A. dorsata was individually extracted using a SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. The almost entire mitochondrial genome (except for tRNASer and D-loop) of Apis dorsata was 15,275 bp and deposited in the GenBank under the accession number KC294229 in NCBI. Annotation of the complete assembled mitochondrial genome was performed with Dual OrganellarGenoMe Annotator (DOGMA) (Wyman et al. Citation2004). The mitogenome sequenced harbours 13 protein-coding genes (PCGs), 21 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA), and one control region (D-loop). The total base composition was A (42.38%), T (42.27%), G (5.82%), and C (9.53%), suggesting that the percentage of A + T (84.65%) was higher than G + C (15.35%), which is similar, but slightly different from other Apis species.
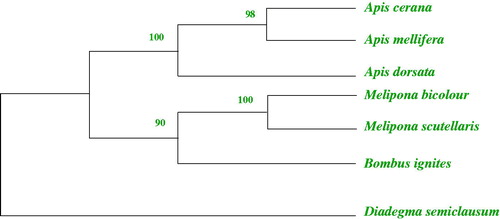
To better understand phylogenetic relationship of the Apis with other closely related species, multiple alignments of seven mitochondrial genomes were performed using Clustal W with the default settings (Thompson et al. Citation1994). The phylogenetic analyses in this study concur with the proposed and widely accepted view of the internal relationships of Apis, A. cerana, and A. mellifera are sister taxa and A. dorsata are more original (Arias and Sheppard Citation2005; Ratnieks Citation2006; Raffiudin and Crozier Citation2007). The consensus tree topology showed Apinae to be a completely monophyletic taxa observed having 1000 bootstrap value (). The complete mitochondrial genomes of six other species are available from GenBank and the science name and accession numbers are as follows: Apis cerana (GQ162109), Apis mellifera ligustica (L06178), Melipona bicolour (NC_004529), Melipona scutellaris (NC_026198), Bombus ignites (DQ870926), and Diadegma semiclausum (EU871947).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arias MC, Sheppard WS. 2005. Phylogenetic relationships of honey bees (Hymenoptera: Apinae: Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 37:25–35.
- Hepburn HR, Radloff SE, Otis GW, Fuchs S, Verma LR, Ken T, Chaiyawong T, Tahmasebi G, Ebadi R, Wongsiri S. 2005. Apis florea: morphometrics, classification and biogeography. Apidologie. 36:359–376.
- Otis GW. 1996. Distributions of recently recognized species of honey bees (Hymenoptera: Apidae; Apis) in Asia. J Kans Entomol Soc. 69:311–333.
- Raffiudin R, Crozier RH. 2007. Phylogenetic analysis of honey bee behavioral evolution. Mol Phylogenet Evol. 43:543–552.
- Ratnieks WFL. 2006. Asian honey bees: biology, conservation and human interactions. Nature. 442:249–249.
- Thapa R, Shrestha R, Manandhar DN, Bista S, Kafle B. 2001. Beekeeping in Nepal. In: Proceedings of the 7th international conference on tropical bees, Chiang Mai, pp. 409–413.
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.