Abstract
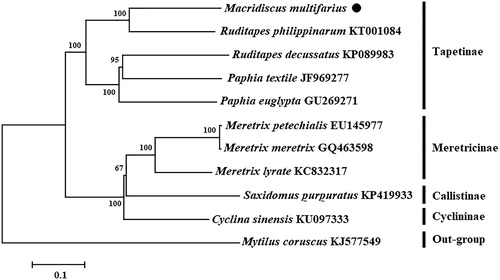
Herein, we first determined the complete mitochondrial genome of Macridiscus multifarius in this study. The circular mitochondrial genome is 20,248 bp in length, consisting of 12 protein-coding genes, 22 tRNA genes and 2 ribosomal RNA genes. The nucleotide composition is 28.6% of A, 39.3% of T, 10.4% of C and 21.7% of G. In 12 protein-coding genes, Cytb, atp6, and nad6 start with ATG, nad1,3 and COX2 with GTT, COX3 and nad4 with ATT, and COX1 and nad5 with ATA. For the stop codon, seven genes with TAA, nad3,4,5 and COX3 with TAG, atp8 ends with AAA. The phylogenetic tree was constructed based on 12 protein-coding genes of 11 species using the Neighbour-joining method, and the result showed that the M. multifarius was closed to the Ruditapes philippinarum. The mitochondrial genome would be helpful for the study of population genetic and phylogenetic analysis of Marcidiscus.
The clam, Macridiscus multifarius (Mollusca, Bivalve, Veneridae) is one of the important bivalve in China coastline aquaculture due to its delicious meat and rich nutrition. It is widely distributed in the northwest Pacific (Zhang et al. Citation2008; Kong et al. Citation2012; Ye et al. Citation2015). Although the M. multifarius is a common species, there is lack of genetic information. It has limited the fisher management and breeding industry to grasp the situation of germplasm resources and select the high-quality parents of M. multifarius. It is the first species in the family Macridiscus for which complete mitochondrial genome has been determined in this study.
The sample of M. multifarius was identified by morphology in Zhejiang province, China (122.2°E, 30.3°N) using BLAST in NCBI database to ensure that the sample did not belong to other species. The total genomic DNA extraction was performed by the salting-out method provided in Aljanabi and Martinez (Citation1997) using the adductor muscle. The total genomic DNA was diluted to a final concentration of 50–60 ng/μl in 1 × TE buffer and stored at 4 °C for further analysis. Twenty pairs of primers were utilized to amplify the complete mitochondrial genome. The products were sequenced in Hangzhou TSINGKE Biotechnology Co. Ltd. (Hangzhou, China). We also sequenced the mitochondrial genome on Illumina Hiseq2500 platform using the total genomic DNA.
The complete mitochondrial genome of M. multifarius is 20,248 bp in length (GenBank accession number: MH932410). The complete mitochondrial genome has 12 protein-coding genes, 22 transfer RNA genes (tRNA), and 2 ribosomal RNA genes (12S rRNA and 16S rRNA). The D-loop region is 1944 bp in length between the COX1 gene and Cytb gene. The nucleotide composition for M. multifarius is 28.6% of A, 39.3 of T, 10.4% of C, and 21.7% of G. In 12 protein-coding genes, Cytb, atp6, and nad6 start with ATG, nad1,3 and COX2 with GTT, COX3 and nad4 with ATT, and COX1 and nad5 with ATA. For the stop codon, seven genes with TAA, nad3,4,5 and COX3 with TAG, atp8 ends with AAA. The 16S rRNA is 1345 bp between the tRNAPro and the atp8 genes, and the 12S rRNA is 1175 bp between the COX3 and tRNALeu genes.
The phylogenetic tree () was constructed based on 12 protein-coding genes of 11 species using the neighbour-joining (NJ) method in the program Phylip (Felsenstein Citation1996). The tree showed that the M. multifarius is closely related to Ruditapes philippinarum and belongs to the family Tapetinae. We believe that this result will further supplement the genome information in mitochondria of the family Maridiscus and facilitate the study on population genetic.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693.
- Felsenstein J. 1989. PHYLIP-phylogeny inference package, Version 3.2[J]. Cladistics-The International J Willi Hennig Society, 5(4):164–166.
- Kong L, Matsukuma A, Hayashi I, Takada Y, Li Q. 2012. Taxonomy of Macridiscus species (Bivalvia: Veneridae) from the western Pacific: insight based on molecular evidence, with description of a new species. J Molluscan Stud. 78:1–11.
- Ye YY, Wu CW, Li JJ. 2015. Genetic population structure of Macridiscus multifarius (Mollusca: Bivalvia) on the basis of mitochondrial markers: strong population structure in a species with a short planktonic larval stage. PloS One. 10:e0146260.
- Zhang J, Xiao G, Chai X. 2008. Primary study on artificial breeding of Gomphina venerformis. J Zhejiang Ocean Univ (Nat Sci). 3:008.