Abstract
The complete chloroplast genome of Allium fistulosum (Bunching onion) was determined. The length of the complete chloroplast genome is 153,164 bp. The whole chloroplast genome consists of 82,237 bp long single copy (LSC) and 17,907 bp small single copy (SSC) regions, separated by a pair of 26,510 bp inverted repeat (IR) regions. The Allium fistulosum chloroplast genome encodes 114 annotated known unique genes including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes.
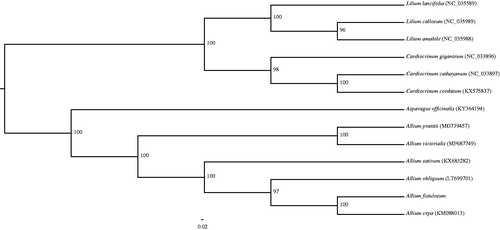
Species of Allium L. section Cepa (Mill.) Prokh. (Amaryllidaceae) are endangered and economical important plants in the world. Allium sect.Cepa consists of 12 species (Gurushidze et al. Citation2007). Two most economically important cultivated species are Allium cepa L. and Allium fistulosum L. A. fistulosum is possibly native to West China and widely cultivated elsewhere (Xu and Kamelin Citation2000). Up to now all phylogenetic analyses on Allium sect. Cepa were based upon a few genes, it is prudent and necessary to examine further evidence before adopting this taxonomic treatment. We assembled the complete cp genome of A. fistulosum to provide genomic and genetic sources for further research on economical important species of section Cepa. The fresh leaves of A. fistulosum were provided by Kunming Botanical Garden, section of Sino-Uzbekistan Global Allium Garden, Yunnan province, China. Voucher specimen was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN, ZY087).Total DNA was extracted from 100 mg of fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). Genomic DNA was fragmented into 500 bp to construct pair-end library. Illumina libraries were prepared according to the manufacturer’s protocol and then sequenced using an Illumina HiSeq 2500 system at BGI (Shenzhen, Guangdong, China). We assembled the cp genome following the approach described in Jin et al. (Citation2018). The plastome of A.cepa was used as reference genome for assembly (Genbank accession: KM088013). The cp genome of A. fistulosum was annotated using Geneious v10.2 (Kearse et al. Citation2012), then start and stop codons and intron/exon boundaries were manual proofread. The annotated chloroplast genome of A. fistulosum has been deposited into the GenBank with the Accession number MH926357. Phylogenetic analysis was performed using RAxML-HPC BlackBox v8.1.24 software (Stamatakis Citation2006) with the GTRGAMMAI model as suggested (RAXML manual). The phylogenetic analysis of 12 chloroplast genomes showed that A. fistulosum is closely related to A. cepa ().
Figure 1. Phylogenetic analysis of A. fistulosum with 12 related species. Numbers in the nodes are the bootstrap values from 1000.
The complete cp genome sequence of A. fistulosum is 153,164 bp and shows a characteristic circular structure, including a pair of IRs (26,510 bp, GC – 42.7% for each) that divide the genome into two single-copy regions (LSC 82,237 bp, GC – 34.6%; SSC 17,907 bp, GC – 29.7%), whereas coding regions (92,124 bp, GC – 39.5%) and non-coding regions (92,124 bp, GC – 32.8%). There are a total of 134 genes in the genome, including 80 protein-coding genes, 30 tRNA genes, four ribosomal RNA genes, and 20 duplicated genes. Among these, 13 genes (atpF, rps16, rpoC1, petB, petD, rpl16, two rpl2, two rps12, two ndhB and ndhA) contain a single intron and two genes (ycf3 an clpP) contain double intron . The two (ycf15 and rps2) genes are putattive pseudogenes in the coplete cp geneome of A. fistulosum. This complete cp genome can be further used for population genomic studies, phylogenetic analyses, genetic engineering studies of genus Allium.
Acknowledgements
The authors are grateful to the opened raw genome data from public database. The authors thank Dr. Yunheng Ji for the help of sequence analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. https://doi.org/10.1101/256479
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gurushidze M, Mashayekhi S, Blattner FR, Friesen N, Fritsch RM. 2007. Phylogenetic relationships of wild and cultivated species of Allium section Cepa inferred by nuclear rDNA ITS sequence analysis. Plant Syst Evol. 269:259–269.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Xu JM, Kamelin RV. 2000. Allium L. In: Wu Z.Y. , Raven P.H.. (eds) Flora of China, Vol. 24. Beijing, St. Louis: Science Press, Missouri Botanical Garden Press; p. 193.
