Abstract
Laurus azorica is an endemic tree species from Azores archipelago. In this study, we generated the plastome sequence as a first step in a global assessment of island endemic Lauraceae. The total chloroplast genome was 153,061 bp in length, with a large single copy (LSC) region of 87,226 bp and a small single copy (SSC) region of 18,033 bp, separated by two inverted repeats (IRs) regions of 23,901 bp. The overall GC content was 37.3%, and 43.1%, 35.4%, and 31.1% in the IRs, LSC, and SSC regions, respectively. It contained 106 genes, including 79 coding genes, 24 tRNA genes, and four rRNA genes. A phylogenetic analysis confirmed that L. azorica was clustered with L. nobilis within the family Lauraceae.
Lauraceae are an ancient family of woody plants found in tropical and subtropical regions, with centers of diversity in South America and Southeast Asia (Chanderbali et al, Citation2001). They comprise more than 3000 species in ∼50 genera worldwide. In addition to high commercial value for edible fruits (e.g. avocado – Persea americana) and spices (e.g. cinnamon – Cinnamomum verum), Lauraceae are also an important structural and evolutionary component of wet tropical forests (Christenhusz and Byng Citation2016).
However, despite their abundance and widespread occurrence, Lauraceae remain poorly known and difficult to identify, primarily due to the absence of clear morphological and molecular diagnostic characters (Werff and Geography Citation2001, Little et al, Citation2009). Plastome sequencing can therefore increase the identification success by providing a comprehensive set of data (Hinsinger and Strijk Citation2017).
Laurus L. is the type genus of the Lauraceae family and holds three species, L. nobilis L. (the true laurel) being used as culinary herb, and two other species, L. azorica and L. novocanariensis, endemic from Azores and Canary Islands, respectively (Córdoba and Medina Citation1976; Barbero et al. Citation1980). Here we report the complete chloroplast sequence of L. azorica to provide resources for delineation of the taxonomical status of this species and improve its in situ conservation.
Laurus azorica is a dioecious tree, endemic to the Azores, present in all islands of the archipelago. It is one of the most frequent species in montane cloud forests of the Azores (Elias et al. Citation2011) but in the past it was probably much more abundant because it was one of the typical laurel forest species (especially in the submontane belt), that dominated the landscape of the archipelago before human settlement (Elias et al. Citation2016). Presently, L. azorica may be found mostly between 100 and 1000 m a.s.l., but it is more abundant between 300 and 600 m a.s.l., and frequent up to 900 m. In the past L. azorica berries were used to produce oil for medicinal purposes and lighting (Dias Citation1946). Although this was not the preferred tree for the purpose, the wood of this species was also used to produce coal (Costa Citation1950). Nevertheless, the main cause for the decline of the species was the replacement of Laurel forests by pastures and plantation forests, and invasion by exotic trees. As L. azorica occurs on the nine islands of the Azores archipelago, we expect its plastome sequence will provide valuable data to further study the population structure among and between islands, and will improve our understanding of the systematics and the evolutionary history of the Lauraceae.
Genomic DNA was extracted from silica-dried leaves of L. azorica collected on the Terceira island (38°45′00.3ʺN 27°19′57.4ʺW), using a modified SDS protocol (Healey et al. Citation2014). Library construction and sequencing were performed by Novogene (Beijing, PR China) on an Illumina Hiseq2500 system, following manufacturer instructions. Vouchers were deposited in the herbarium of the Azores University (AZU), with DNA material stored in the laboratory of the Biodiversity Genomics Team (Guangxi University, Nanning, PR China), under accession BGT3615. We performed a de novo assembly using ORG.asm (http://phythonhosted.orgorg.asm/), and annotated the assembled plastome using cpGAVAS (Schenk et al. Citation2012), followed by manual adjustments in Geneious (R9, Biomatters, www.geneious.com).
The plastome of L. azorica was 153,011 bp in length. It contains a large single-copy (LSC) region (93,511 bp), a small single-copy (SSC) region (18,492 bp), and two inverted repeats (IRs) (41,008 bp). 179 genes, with 80 coding regions, 28 ribosomal RNA genes and 95 tRNA genes were annotated. The overall GC content of the cp genome was 39.1%, and 44.1%, 37.9%, 33.9% in the IRs, LSC and SSC regions, respectively. The plastome sequence was deposited in Genbank (MK041220).
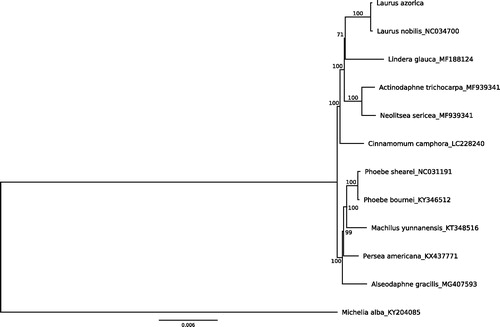
Ten other Lauraceae plastomes were retrieved from GenBank, and used to reconstruct a phylogenetic tree using PhyML (ref). The ML tree highlighted both the close relationship of L. azorica with L. nobilis and of the Laurus spp. with Lindera glauca (). Our results also confirmed the split of the family in two main groups. The length of the branch leading to the genus Laurus indicates either an old divergence from the rest of the family, or a possible increase in the plastome substitution rate as Laurus adapted to the Mediterranean climate.
Acknowledgements
We would like to acknowledge Fernando Pereira for sampling the material of Laurus azorica.
Additional information
Funding
References
- Barbero M, Benabid A, Peyre C, Quézel PJADJBDM. 1980. Sur la presence au Maroc de "Laurus azorica" (Seub.). Franco. 37:467–472.
- Chanderbali AS, Werff HVD, Renner SS. 2001. Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Missouri Bot Gard. 88:104–134.
- Christenhusz MJM, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261:201–217.
- Córdoba LCYFD, Medina FO. 1976. Estudio sobre la vegetación y flora forestal de las Canarias occidentales.(1): p1-6. Excmo, Cabildo Insular.
- Costa C. 1950. Arvoredos dos Açores: algumas achegas para a sua história. Comissão Reguladora Dos Cereais Do Arquipélago Dos Açores. 11:45–60.
- Dias UM. 1946. Ponta Delgada: descrição de quando foi lugar e vila e de cidade: escorço histórico. Tipografia A Crença. Ponta Delgada, Açores.
- Elias RB, Dias E, Pereira F. 2011. Disturbance, regeneration and the spatial pattern of tree species in Azorean mountain forests. Commun Ecol. 12:23–30.
- Elias RB, Gil A, Silva L, Fernandez-Palacios JM, Azevedo EB, Reis F. 2016. Natural zonal vegetation of the Azores Islands: characterization and potential distribution. Phytocoenologia. 46:107–123.
- Healey A, Furtado A, Cooper T, Henry RJJPM. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 10:1–8.
- Hinsinger DD, Strijk JSJ. 2017. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Little SA, Stockey RA, Penner B. 2009. Anatomy and development of fruits of Lauraceae from the Middle Eocene Princeton Chert. Am J Bot. 96:637–651.
- Schenk L, Schnitzer S, Adolph H, Holzhausen J, Matheis E, Kuhlmey AJBG. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–715.
- Van Der Werff HJ. 2001. An annotated key to the genera of Lauraceae in the flora Malesiana region. Blumea 46:125–140.