Abstract
The Sarcophagidae family is one of the most important in forensic entomology. Its richest genus, Oxysarcodexia, is well-distributed in Brazil and Latin America and includes species that can be used in forensic investigations to estimate the Post Mortem Interval (PMI). In this communication, we present the mitochondrial genome (mtDNA) of four Oxysarcodexia species: O. avuncula (one specimen), O. terminalis (one specimen), O. thornax (three specimens), and O. varia (one specimen). These mitochondrial genomes (mtDNA) range from 14,998 to 15,613 bp and have 22 tRNA genes, 13 protein-coding genes (PCG), and 2 rRNAs distributed along both strands. The mitogenomes from Oxysarcodexia showed in the present work represents an important contribution to the knowledge regarding the Sarcophagidae phylogenetic structure and is an important source of information for the development of novel DNA markers for forensic identification.
The genus Oxysarcodexia is one of the most diverse in the Sarcophagidae family, with 86 species described (Pape Citation1996), and is mainly observed in Latin America (Madeira et al. Citation2016). Species of this genus are frequently used to estimate the Post Mortem Interval (PMI) in forensic investigations (Madeira et al. Citation2016). Nevertheless, species identification depends on observation of male genitalia, thus hindering the identification of larvae and female individuals (Silva and Mello-Patiu Citation2008) and also of dandified specimens from this genus. We show novel complete mtDNA sequences of flesh fly species of forensic importance. Our data expand the knowledge on the molecular database for identification of these species.
In this work, we collected specimens from different places in Brazil: O. avuncula (voucher NT5 – 23°18'27" S, 47°07'59" W); O. terminalis (voucher 200 – 17°43'17" S, 48°09'40" W); O. varia (voucher 469 – 22°51'12" S, 46°19'18" W); and the three specimens of O. thornax were in (voucher 297 – 19°44'59" S, 47°56'02" W; voucher 433 – 31°26'49" S, 53°06'02" W; and voucher 506 – 23°10'13" S, 46°53'53" W). Each specimen was identified following taxonomic keys (Carvalho and Mello-Patiu Citation2008; Silva and Mello-Patiu Citation2008) and preserved in 70% ethanol. The mtDNA was extracted from the thorax, legs, and wings (Françoso et al. Citation2016), and the head and abdomen were deposited in the database of UNICAMP according to the voucher codes mentioned above. The mtDNA library was generated using the Nextera XT kit, according to the manufacturer’s instructions and sequenced using a paired-end strategy 2x250 on the MiSeq platform (Illumina, CA). The genomes were constructed by mapping the reads against the Sarcophagidae mitogenomes available on the GeneBank database, followed by de novo assembly of the mapped reads using the CLC Genomics Workbench. The mitogenomes were annotated using the MITOS WebServer (Bernt et al. Citation2013) and manually verified using the NCBI database.
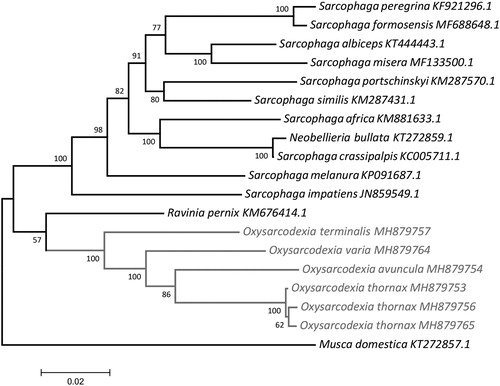
The complete mtDNA sequence of O. avuncula (GenBank, MH879754) was 14,998 bp long and presented 24% of GC. Oxysarcodexia terminalis (GenBank, MH879757) mtDNA was 15,613 bp long and showed 24.3% of GC. Oxysarcodexia thornax sequences were 15,237 bp (voucher – GenBank, 297 – MH879756, and 433 – MH879753) and 15,238 bp (voucher 506 – MH879765) long, with 23.8% of GC. The mtDNA sequence of O. varia (MH879764) was 15,261 bp long and presented 24% of GC. The annotation of these mtDNAs revealed 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a noncoding control region (D-loop) located between the 12S rRNA and tRNAIle. While most of the genes are encoded in the heavy strand, the light strand codifies five PCGs, 8 of the 22 tRNAs, and the 2 rRNAs. Five PCGs have the T–– stop codon completed to TAA by post-transcriptional polyadenylation (Ojala et al. Citation1981). The PCG sequences commonly start with an ATT, ATA, or ATG codon (12 PCGs), but COX1 starts with CAA. Phylogenetic analysis was done with the whole mtDNA of these species and more 13 mtDNA available in GenBank ().
Figure 1. Molecular phylogenetic inferences of Oxysarcodexia mitogenomes. The phylogenetic analysis was based on the maximum likelihood (ML) with the nearest-neighbor-interchange heuristic method using MEGA 7 software (Kumar et al. Citation2016). The D-loop region was excluded from this analysis due to its high degree of variability (Gonder et al. Citation2007). The tree showed a clearly separated branch for the species and suggests that the Oxysarcodexia clade is monophyletic and well-supported by the branches and is closer to the genus Ravina. The individual differences among O. thornax specimens may be due to the geographical separation of the populations, as the individuals MH879756 and MH879765, collected from locations separated by approximately 100 km, showed higher similarity to each other in comparison with MH879753, which was collected approximately 1300 and 1400 km away from MH879765 and MH879756, respectively. The Oxysarcodexia specimens are highlighted in grey. The mitogenomes for comparison were obtained from the NCBI database: Musca domestica (KT272857.1) – used as outgroup, Sarcophaga impatiens (JN859549.1), Sarcophaga melanura (KP091687.1), Neobellieria bullata (KT272859.1), Sarcophaga crassipalpis (KC005711.1), Sarcophaga peregrina (KF921296.1), Sarcophaga formosensis (MF688648.1), Sarcophaga africa (KM881633.1), Sarcophaga portschinskyi (KM287570.1), Sarcophaga similis (KM287431.1), Sarcophaga albiceps (KT444443.1), Sarcophaga misera (MF133500.1), Ravinia pernix (KM676414.1).
Acknowledgments
We thank the Universidade de Lisboa and the Universidade Federal de Minas Gerais (UFMG) for the support during the project execution.
Disclosure statement
The authors report no potential conflict of interest.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Carvalho CJB, Mello-Patiu CA. 2008. Key to the adults of the most common forensic species of diptera in South America. Rev Bras Entomol. 52:390–406.
- Françoso E, Gomes F, Arias MC. 2016. A protocol for isolating insect mitochondrial genomes: a case study of NUMT in Melipona flavolineata (Hymenoptera: Apidae). Mitochondrial DNA. 27:2401–2404.
- Gonder MK, Mortensen HM, Reed FA, Sousa A, Tishkoff SA. 2007. Whole-MtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 24:757–768.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol Evol Genetic Anal. 33:1870–1874.
- Madeira T, Souza CM, Cordeiro J, Thyssen PJ. 2016. The use of DNA barcode for identifying species of Oxysarcodexia townsend (Diptera: Sarcophagidae): a preliminary survey. Acta Tropica. 161:73–78.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470.
- Pape T. 1996. Catalogue of the Sarcophagidae of the World (Insecta: Diptera). Memoirs of Entomology International. 8:1–558. ISSN: 1083-6284.
- Silva KP, Mello-Patiu C. 2008. Morfologia comparada da terminalia masculina de quatro espécies de Oxysarcodexia townsend, 1917 (Diptera, Sarcophagidae). Arq Mus Nac Rio J. 66:363–372.
