Abstract
It is first reported on complete chloroplast genome of Asplenium nidus, an economically important fern. The genome is 156,173 bp in length with 40.8% GC content, comprising a pair of inverted repeats (22,834 bp for each) separated by a large single copy (89,014 bp) and a small single copy (21,491 bp) region. It encodes 130 genes, including 88 protein-coding genes, 33 tRNA genes, eight rRNA genes, and one pseudogene. The maximum-likelihood tree shows that A. nidus is sister to A. prolongatum with 100% support value. The complete chloroplast genome of A. nidus provides powerful valuable molecular data to address classification issues of Asplenium.
Asplenium nidus L., commonly known as bird's nest fern, is an epiphytic fern in Aspleniaceae (Lin and Viane Citation2013). It is clustered on tree trunks or rocks in rain forests at an elevation of 100–1900 m. Fronds are large simple with bright green, forming a rosette of leaves at the top of rhizome (Jia et al. Citation2016). Veins are straight with a slight angle to black midrib and usually once-forked (Lin and Viane Citation2013). Asplenium nidus is mainly distributed in tropics and subtropics of the Old World (Dong Citation2011). Except as a key ecological role (Karasawa and Hijii Citation2006; Ellwood et al. Citation2009), the fern also has important economic values as edible vegetable, folk medicine for asthma, and ornamental plant (Jia et al. Citation2016). Asplenium is a very large genus with more than 700 species. Some taxa resembling A. nidus have been recognized as a separate section Thamnopteris (Lin and Viane Citation2013). However, the treatment is not supported by recent molecular studies (Ohlsen et al. Citation2014). As the first member in this section (Linnaeus Citation1753), A. nidus has been found to be polyphyletic to other species (Ohlsen et al. Citation2014). The taxon delineation and cryptic species identification are to be further explored (Yatabe et al. Citation2009; Dong Citation2011). Hence, acquirement of whole chloroplast (cp) genome sequence of A. nidus will contribute to revise the taxonomy of this difficult section and lay solid foundations for further phylogenomic investigation.
Fresh leaves of A.nidus were collected from South China Botanical Garden, Chinese Academy of Science (23°11′3.56″N, 113°21′43.28″E). The voucher specimen was kept by the Herbarium of Sun Yat-sen University (SYS; SS Liu 2020). Total genomic DNA was extracted using the Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). We constructed genomic library and sequenced with pair-end (2 × 300 bp) in Illumina Hiseq 2500 platform (IIIumina Inc., San Diego, CA, USA). After quality trimming of raw reads with Trimmomatic v0.32 (Bolger et al. Citation2014), we obtained a total of 11,498,218 clean reads, which were further assembled into complete chloroplast genome using Velvet v1.2.07 (Zerbino and Birney Citation2008). Plastome annotation was implemented by the Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and validated with BLAST searches. The tRNAs were further confirmed using tRNAscan-SE 1.21 (Schattner et al. Citation2005). A maximum likelihood (ML) analysis was performed based on the complete chloroplast genomes of 13 ferns including Alsophila podophylla as outgroup using RaxMLv.8.0 (Stamatakis Citation2014) with 1000 bootstrap replicates.
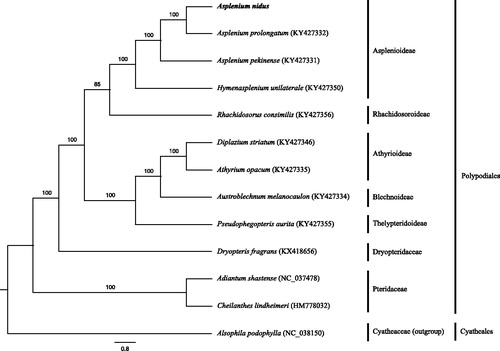
The complete chloroplast genome was 156,173 bp in length with 40.8% GC content, including a pair of inverted repeats (22,834 bp for each), a large single copy (89,014 bp) and a small single copy (21,491 bp) regions (GenBank accession number: MK002975). It encodes 130 genes, involving in 88 protein-coding genes (PCG), 33 tRNA genes, eight rRNA genes, and one pseudogene. Fourteen genes are duplicated, including four PCGs (rps12, rps7, psbA, and ycf2), six tRNA genes (trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, trnT-UGU, and trnR-ACG) and four rRNA genes (rrn4.5, rrn5, rrn23, and rrn16). Three genes (ycf3, clpP, and rps12) contain two introns. ML tree strongly shows that A. nidus is sister to A. prolongatum, which further forms a monophyletic clade to A. pekinense with 100% support value (). The complete chloroplast genome of A. nidus provides powerful valuable molecular data to address classification issues of Asplenium.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Dong SY. 2011. Taxonomic studies on Asplenium sect. Thamnopteris (Aspleniaceae) I: cytological observations. Am Fern J. 101:156–171.
- Ellwood MDF, Manica A, Foster WA. 2009. Stochastic and deterministic processes jointly structure tropical arthropod communities. Ecol Lett. 12:277–284.
- Jia X, Deng Y, Sun X, Liang L, Su J. 2016. De novo assembly of the transcriptome of Neottopteris nidus using Illumina paired-end sequencing and development of EST-SSR markers. Mol Breed. 36:94.
- Karasawa S, Hijii N. 2006. Does the existence of bird's nest ferns enhance the diversity of oribatid (Acari: Oribatida) communities in a subtropical forest? Biodivers Conserv. 15:4533–4553.
- Lin YX, Viane R. 2013. Aspleniaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 267–316.
- Linnaeus C. 1753. Species plantarum. Stockholm: Laurentius Salvius.
- Ohlsen DJ, Perrie LR, Shepherd LD, Brownsey PJ, Bayly MJ. 2014. Phylogeny of the fern family Aspleniaceae in Australasia and the south-western Pacific. Aust Systematic Bot. 27:355–371.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yatabe Y, Shinohara W, Matsumoto S, Murakami N. 2009. Patterns of hybrid formation among cryptic species of bird-nest fern, Asplenium nidus complex (Aspleniaceae), in West Malesia. Bot J Linn Soc. 160:42–63.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.