?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Anemarrhena asphodeloides represents the only species in monotypic genus Anemarrhena. Its rhizome is called Zhi Mu and has been used in traditional medicine in China for more than 2000 years. We sequenced the complete chloroplast (CP) genome of this plant. The CP genome of A. asphodeloides is 157,734 bp in length, containing a pair of 53,313 bp inverted repeat regions (IRs) separated by one large and one small single copy region (LSC and SSC) of 85,851 and 18,570 bp, respectively. The over-all AT content of the CP genome is 62.15%. The phylogenetic analysis strongly supported that A. asphodeloides comprises a monophyly, and it is affiliated to Subfam. Agavoideae (Asparagaceae). The results will be useful for conservation, phylogenetics, and identification of this species.
Anemarrhena asphodeloides Bunge is one of the important medicinal plants, which has been used for over 2000 years in China. It has the effects of clearing away heat and reducing fire, nourishing yin, and moistening dryness (Han and Park Citation1997; Chinese Pharmacopoeia Commission Citation2015). Presently, the wild A. asphodeloides has become quite rare in China as a result of over-exploitation and the deterioration of natural conditions (Sun et al. Citation2008). A. asphodeloides is the only species in genus Anemarrhena, a taxonomically controversial group, whose phylogenetic position has frequently changed during the past 40 years (Delectis Florae Reipublicae Popularis Sinecae Citation1980; APG Citation2016). In this study, we sequenced the complete chloroplast genome of A. asphodeloides, in order to give a phylogenomic insight to systematic position of the genus Anemarrhena and provide genetic information for conservation of this species.
Fresh leaves of A. asphodeloides were collected from Pingshan County, Hebei Province in China (Latitude: 38°130.09″N, Longitude: 114°6′13.69″E). The voucher specimen (Chao Zhi 147311) was identified by Prof. Zhi Chao and deposited in the herbarium of the School of Traditional Chinese Medicine, Southern Medical University. The genomic DNA was extracted from 100 mg of fresh leaves with the modified CTAB method (Yang et al. Citation2014). Sequencing was performed on an Illumina HiSeq 4000 in high output mode with 2 × 125 bp paired-end reads at Beijing Genomics Institute (BGI, Shenzhen, China). The CP genome of A. asphodeloides was assembled by SOAPendvo2.04 (Luo et al. Citation2012) with Agave attenuata as a reference (Accession No. NC032696). The Dual Organellar Genome Annotator (DOGMA) software (Wyman et al. Citation2004) was used for genome annotation. The annotated sequence had been deposited in GenBank (Accession No. MH669277).
The complete CP genome of A. asphodeloides was a double-stranded circular DNA of 157,734 bp in length. Its quadripartite structure was composed of two inverted repeated regions (IRa and IRb) of 53,313 bp, separated by a large single-copy (LSC) region of 85,851 bp and a small single-copy (SSC) region of 18,570 bp. The complete CP genome contains a total of 112 genes, including 88 protein-coding genes, 20 tRNA genes, and 4 rRNA genes. Eighteen genes were duplicated in the IR regions. Fifteen genes contain one intron, while three genes have two introns. The over-all A-T content of the chloroplast genome is 62.15%.
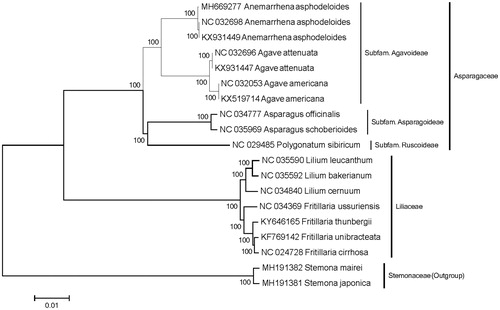
A phylogenetic analysis was performed based on 19 complete CP genome, to reveal the phylogenetic position of genus Anemarrhena. All the CP genome sequences were aligned with MAFFT (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was inferred in MEGA 6.0 based on GTR + R + I model using 1000 bootstrap replicates (Tamura et al. Citation2013). The reconstructed phylogenetic tree showed that genus Anemarrhena was closely related to genus Agave and was affiliated to Subfam. Agavoideae (Asparagaceae), which is consistent with the APG IV. (Angiosperm Phylogeny Group Citation2016) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Chinese Pharmacopoeia Commission. 2015. Pharmacopoeia of the People's Republic of China, 2015 Edition. Bejing: China Medical Science and Technology Press. p. 212.
- Delectis Florae Reipublicae Popularis Sinecae. 1980. Flora Reipublicae Popularis Sinecae, Tomus 14. Beijing, China: Science Press. p. 40.
- Han SH, Park SI. 1997. Classification of Korean native Anemarrhena asphodeloides Bunge by cluster analysis. Korean J Med Crop Sci. 5:266–275.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. 2012. SOAPdenono2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18–23.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvementsin performance and usability. Mol Biol Evol. 30:772–780.
- Sun XM, Chen QL, Wang WQ, Ma CH, Zhang Y. 2008. Investigation of Anemarrhena asphodeloides Bge. wild resources and its germplasm analysis. Lishizhen Med Mater Med Res. 19:2091–2092.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 14:1024–1031.