Abstract
This study presents the first mitogenome of Daphnia laevis, a species that plays an important ecological role in Brazilian aquatic environments. The mitochondrion is 15,246 bp in size, with 13 protein-coding genes, 22 tRNAs genes, two rRNAs and d-loop region. Fourteen overlaps were found, a total of 98 bp. The percentages of A, T, C and G were 35.0%, 33.6%, 18.0%, and 13.4%, respectively, with A + T 68.6%. A cluster analysis confirmed that Daphnia laevis is close to other Daphnia species.
Cladocerans represent a key group in freshwater environments. They present a wide geographic distribution, with Daphniidae predominant in Brazil (Rocha and Güntzel Citation1999). Daphnia laevis is considered endemic to the American continent, being registered in North (Taylor et al. Citation1998), Central (Collado et al. Citation1984; Lalana et al. Citation2005) and South America (Paggi Citation1977), including Brazil (Matsumura-Tundisi Citation1984; Rocha et al. Citation2011). In addition to its ecological relevance, the species has demonstrated sensitivity to toxic compounds (Arauco et al. Citation2005; Rietzler et al. Citation2017) and has potential as a model organism in Ecotoxicology.
Specimens of D. laevis were collected in the water column, using zooplankton nets in a lake adjacent to the Parque Estadual do Rio Doce (PERD), southeastern Brazil (Jacaré Lake, 19S48′38″, 42W38′55’″). After sampling, females were isolated and individually cultured in the laboratory.
Mitochondrial DNA extraction followed the methodology of Françoso et al. (Citation2016). The paired-end libraries were built by applying the Nextera XT DNA kit from Illumina (Illumina Citation2017) and the mitogenome was sequenced using the Illumina MiSeq sequencing platform – Next Generation Sequencing (NGS). The sequenced result was imported to Geneious 10.2 to be trimmed with the default setting. The COI sequence of D. laevis (GenBank ID) was used as seed to assemble the mitogenome. The annotation was performed using MITOS 2 Webserve (Bernt et al. Citation2013) and similarities were verified by the NCBI database. The evolutionary history was inferred using the Neighbor-Joining method and the evolutionary distances were computed using the Kimura 2 parameter method conducted in MEGA7.
The D. laevis mitogenome (MK059395) is 15,246 bp in length, 22 tRNAs genes, typical arthropod tRNAs (tRNASer2, tRNALeu1, tRNAVal, tRNAIle, tRNAGln, tRNAMet, tRNATrp, tRNACys, tRNATyr, tRNALeu2, tRNALys, tRNAAsp, tRNAGly, tRNAAla, tRNAArg, tRNAAsn, tRNASer1, tRNAGlu, tRNAPhe, tRNAHis, tRNAThr, tRNAPro), 13 protein-coding genes (COX1, COX2, COX3, CYTB, ATP8, ATP6, ND1, ND2, ND3, ND4, ND4L, ND5, ND6), two rRNAs (16S rRNA and 12S rRNA) separated by tRNAVal, as similarly found in D. pulex and D. carinata (Geng et al. Citation2016a, 2016Citationb) and d-loop region (544 bp). Among all 37 genes, 23 were encoded on the H-strand. Fourteen overlaps were found between adjacent genes, involved a total of 98 bp, but D. galeata occurs 11 times (30 bp) (Tokishita et al. Citation2017) and D. pulex, six times (26 bp) (Crease Citation1999). The percentage of A, T, C and G were 35.0%, 33.6%, 18.0% and 13.4%, respectively, with A + T (68.6%) similar to D. magna 67.1% (Ruixue et al. Citation2016), D. carinata 70.3% (Geng et al. Citation2016a) and D. pulex 64.5% (Geng et al. Citation2016b), 62.3% (Crease Citation1999). A cluster analysis showed that D. laevis is close to other Daphnia species and Cladocera, grouped. Tokishita et al. (Citation2017) using a phylogram of total protein-coding sequences also located D. carinata close to D. pulex. The Isopoda (outgroup) was grouped separately ().
Figure 1. Evolutionary relationships of taxa obtained by the Neighbor-Joining method based on total genome and excluding d-loop. Ligia oceanica, Gyge ovalis, Cymothoa indica and Limnoria quadripunctata were used as outgroups.
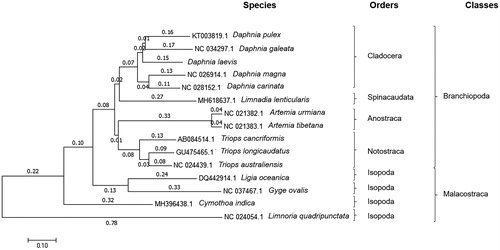
Considering that most Brazilian species of Cladocera are still unknown from the genetic point of view, it is expected that the mitogenome of D. laevis reported here can provide a stimulus for more molecular studies.
Acknowledgements
The authors thank the Universidade Federal de Minas Gerais (UFMG) and the Long-Term Study Program (PELD, site 4)/National Council for Scientific and Technological Development (CNPq) for logistic support.
Disclosure statement
The authors report no potential conflict of interest.
Additional information
Funding
References
- Arauco LRR, Cruz C, Neto JM. 2005. Efeito da presença de sedimento na toxicidade aguda do sulfato de cobre e do triclorfon para três espécies de Daphnia. Pesticidas (UFPR). 15:55–64.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Collado C, Fernando CCH, Sephton D. 1984. The freshwater zooplankton of Central America and the Caribbean. Hydrobiologia. 113:105–119.
- Crease TJ. 1999. The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea). Gene. 233:89–99.
- Françoso E, Gomes F, Arias MC. 2016. A protocol for isolating insect mitochondrial genomes: a case study of NUMT in Melipona flavolineata (Hymenoptera: Apidae). Mitochondrial DNA. A DNA Mapp Seq Anal. 27:2401–2404.
- Geng XX, Cheng R, Deng D, Zhang H. 2016. The complete mitochondrial DNA genome of Chinese Daphnia carinata (Clasocera: Daphniidae). Mitochondrial DNA Part B. 1:323–325.
- Geng XX, Zhang L, Xu M, Deng DG, Zhang HJ. 2016b. The complete mitochondrial genome of the Chinese Daphnia pulex (Cladocera, Daphniidae). ZooKeys. 615:47–60.
- Illumina, Nextera XT DNA Library Prep Reference Guide. Document. 2017. [accessed 2017 August 21] https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-03.pdf
- Lalana R, Ortiz M, Varela C. 2005. Primera adición a la lista de los crustáceos no decápodos de Cuba. Revista de Biología. 19:50–56.
- Matsumura-Tundisi T. 1984. Ocurrence of species of the genus Daphnia in Brazil. Hydrobiologia. 112:161–165.
- Paggi JC. 1977. Aportes al conocimiento de la fauna Argentina de cladoceros 1 Sobre Daphnia laevis Birge 1878. Neotropica. 69:33–37.
- Rietzler AC, Rocha O, Fonseca AL, Ribeiro MM, Matos MF. 2017. Sensitivity of tropical cladocerans and chironomids to toxicants and their potential for routine use in toxicity tests. In: Araújo CVM, Shinn C, editors. Environment, science, engineering and technology: ecotoxicology in Latin America. 1st ed. New York: Nova Publishers; p. 87–103.
- Rocha O, Güntzel A. 1999. Crustáceos Branquiópodos. In: Joly CA, Bicudo CEM, editors. Biodiversidade do Estado de São Paulo, Brasil: síntese do conhecimento ao final do século XX. São Paulo: FAPESP; p. 109–120.
- Rocha O, Santos-Wisniewski MJ, Matsumura-Tundisi T. 2011. Checklist de Cladocera de água doce do Estado de São Paulo. Biota Neotrop. 11:571–592.
- Ruixue C, Bin D, Yaling W, Xuexia G, Jun L, Xu Z, Shuying P, Daogui D, Haijun Z. 2016. Complete mitochondrial genome sequence of Daphnia magna (Crustacea: Cladocera) from Huaihe in China. J. Lake Sci. 28:414–420.
- Taylor DJ, Finston TL, Hebert PDN. 1998. Biogeography of a widespread freshwater crustacean: pseudocongruence and cryptic endemism in the North American Daphnia laevis complex. Evolution. 52:1648–1670.
- Tokishita S, Shibuya H, Kobayashi T, Sakamoto M, Ha JY, Yokobori SI, Yamagata H, Hanazato T. 2017. Diversification of mitochondrial genome of Daphnia galeata (Cladocera, Crustacea): Comparison with phylogenetic consideration of the complete sequences of clones isolated from five lakes in Japan. Gene. 611:38–46.
