Abstract
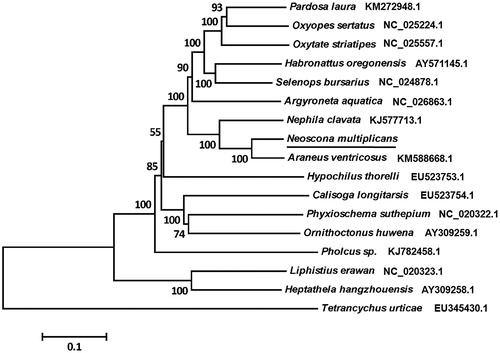
The complete mitochondrial genome of an orb-weaving spider Neoscona multiplicans was determined. The circular mitogenome is 14,074 bp in length (GenBank accession number MK052682), consisting of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA genes, and an A + T-rich region. The gene arrangement and orientation of N. multiplicans are identical to those found in other known spiders. The nucleotide composition is significantly biased toward A + T content at 74.82% (A: 35.44%; C: 9.53%; G: 15.65%; T: 39.38%). ATT, ATA, TTA, TTG were initiation codons and TAA, TAG, T were termination codons. Six intergenic regions and 24 reading frame overlaps were found in the mitogenome of N. multiplicans. The A + T-rich is 483 bp with an A + T content of 77.23%. Eleven tRNAs lack the potential to form the cloverleaf-shaped secondary structure. Phylogenetic tree based on 13 PCGs showed that N. multiplicans is closely related to Araneus ventricosus, which is consistent with the conventional classification.
The orb-weaving spider belongs to the family Araneidae, the third largest family in Araneae, comprising more than 3000 described species in 169 genera (Platnick Citation2015). Most of these spiders are important predators of economically agricultural pests, and play crucial roles in biological control of farmland (Clausen Citation1986; Pearce and Venier Citation2006). The species, Neoscona multiplicans (Chamberlin, 1924) (Araneae: Araneidae), is widely distributed in Japan, Korea and China. In this study, adult individuals of N. multiplicans were collected from Libo county (N25tyted E107tyteda Guizhou Province, China on June 2018. After morphological identification, the specimens were persevered in the spider specimen room of Guiyang University with an accession number GYU-GZML-11.
The complete mitogenome of N. multiplicans is a double-stranded circular molecule of 14,074 bp in length and has been deposited in the GenBank under accession number: MK052682. It harbours 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rrnL and rrnS), and an A + T-rich region, as observed in other metazoan mitogenomes (Boore Citation1999). The gene arrangement and orientation of N. multiplicans are identical to those found in previously determined spider mitogenomes (Li et al. Citation2016; Wang et al. Citation2016). Twenty-two genes were transcribed on the major strand (J-strand), whereas the others were oriented on the minor strand (N-strand). The nucleotide composition of N. multiplicans mitogenome is significantly biased toward A + T content at 74.82% (A: 35.44%; C: 9.53%; G: 15.65%; T: 39.38%). The AT-skew and GC-skew of this genome were −0.053 and 0.243, respectively. The 13 identified PCGs vary in length from 159 to 1641 bp. The cox1 initiated with TTA as the start codon, cox2, cox3, and nad6 began with TTG, nad2, atp8, nad3, nad5, nad4L, and cob started with ATT, and remaining three PCGs started with ATA. Twelve PCGs terminated with the conventional stop codons TAA (cox1, cox2, cox3, atp6, atp8, nad4, nad4L, nad5, and nad6), or TAG (nad1, nad2, and cob), the nad3 used a single T as stop codon.
Gene overlaps in N. multiplicans mitogenome were present at 24 gene junctions and involved a total of 213 bp. The longest overlap is 28 bp in length and located between nad3 and trnL2. There are six intergenic spacer sequences in a total of 28 bp with length varying from 3 to 6 bp and the largest intergenic spacer is situated between trnN and trnA. The length of 22 tRNAs ranged from 51 bp (trnE) to 67 bp (trnL2), and nine of them were encoded on the N-strand. The A + T content of these tRNAs ranged from 66.15% (trnR) to 84.13% (trnG). Eleven tRNAs lack the potential to form the cloverleaf-shaped secondary structure. Eight of them (trnM, trnW, trnK, trnE, trnF, trnH, trnT, and trnL1) lost the TΨC-stem, three tRNAs (trnA, trnS1 and trnS2) lost the dihydrouracil (DHU) arm. The rrnL gene was located between trnL1 and trnV, and was 1025 bp in length, while the rrnS gene resided between trnV and trnQ, with a length of 694 bp. The major A + T-rich region was located between trnQ and trnM genes with a length of 483 bp, and the A + T content was 77.23%.
Using the sequences of 13 PCGs from the mitogenomes of N. multiplicans and 15 other spiders, the neighbour-joining method was used to reveal the phylogenetic relationship among these species. The result showed that N. multiplicans is closely related to Araneus ventricosus (), which is consistent with the conventional classification.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Clausen HIS. 1986. The use of spiders (Araneae) as ecological indicators. Bull Brit Arachnol Soc. 7:83–86.
- Li C, Wang ZL, Fang WY, Yu XP. 2016. The complete mitochondrial genomes of two orb-weaving spider Cyrtarachne nagasakiensis (Strand, 1918) and Hypsosinga pygmaea (Sundevall, 1831) (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2811–2812.
- Pearce JL, Venier LA. 2006. The use of ground beetles (Coleoptera: Carabidae) and spiders (Araneae) as bioindicators of sustainable forest management: a review. Ecol Indic. 6:780–793.
- Platnick NI. 2015. The world spider catalog, Version 13.5. American Museum of Natural History. Available at: http://research.amnh.org/iz/spides/catalog/.
- Wang ZL, Li C, Fang WY, Yu XP. 2016. The complete mitochondrial genome of orb-weaving spider Araneus ventricosus (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1926–1927.