Abstract
Plecoptera (stoneflies) are an anciently derived order of freshwater insects with two divergent suborders Antarctoperlaria in the southern hemisphere and Arctoperlaria primarily in the northern hemisphere. In this paper, we present the first published mitochondrial genome of an Antarctoperlarian stonefly: the New Zealand stonefly Zelandoperla fenestrata. The Z. fenestrata mitochondrial genome is 16,385 bp, with the typical insect mitogenome complement of 13 protein-coding genes, 22 tRNAs, and two rRNA genes, along with a A + T rich control region. This mitogenome will be of interest to the study of evolutionary divergence within Polyneoptera, and will aid in phylogeographic studies of this species.
Keywords:
Introduction
Stoneflies (order: Plecoptera) are a widespread order of winged freshwater insects, with fossils dating back to the early Permian (Sinichenkova Citation1997; Zwick Citation2000; Bethoux et al. Citation2011; Cui et al. Citation2016). Plecoptera is divided into two sub-orders: Antarctoperlaria in the southern hemisphere and Arctoperlaria, primarily in the northern hemisphere. These two sub-orders diverged around 121 Ma (McCulloch et al. Citation2016). Currently, there are twenty-three published mitochondrial genomes of stoneflies, all from Arctoperlaria.
The stonefly Zelandoperla fenestrata species group (Z. fenestrata, Z. pennulata, Z. tillyardi) is widely distributed across New Zealand (McLellan Citation1977, Citation1999). Because of significant population genetic structure, and the presence of multiple non-dispersive flightless forms on multiple mountain ranges, understanding the phylogeography and evolutionary history of this species is of great interest (McCulloch et al. Citation2009; Dussex et al. Citation2016; Veale et al. Citation2018).
Methods
Library preparation and Illumina sequencing
A single Z. fenestrata nymph identified based on the key in (McLellan Citation1999) was collected from an upper tributary of the Logan Burn stream in Central Otago, New Zealand (45.46884S, 169.96146E) elevation 1007 m, and frozen from live at −70 °C until DNA extraction. We then dissected out the head and forelegs and used a Qiagen DNeasy blood and tissue kit to extract the DNA. We then created a 350 bp paired-end genomic library using an Illumina TruSeq Nano DNA Neoprep, with an average insert size of 607 bp. This was sequenced using an Illumina Hi-Seq with 125 bp reads. DNA remains stored at the University of Otago Zoology Department.
Sequence assembly and analysis
We used CUTADAPT (Martin Citation2011) to remove adapter sequences and trim sequences (cut off =30, minimum length =50). We then used MITOBIM v1.8 (Hahn et al. Citation2013) to extract and align mitochondrial sequences using a COI barcode sequence as a seed (Genbank Accession GQ414619.1). The mitogenome sequence was annotated using the MITOS web server (Bernt et al. Citation2013). We aligned the sequence with all published Plecoptera mitochondrial genomes, along with the mayfly Ephemera orientalis as an outgroup in Geneious® 9.1.8. (Kearse et al. Citation2012) then created a Baysian phylogenetic tree using MrBayes (2,500,000 chain length, 250 sampling interval, 10% burn-in, using an HKY + G + I mutational model (as determined in Topali2 Milne et al. Citation2009).
Results
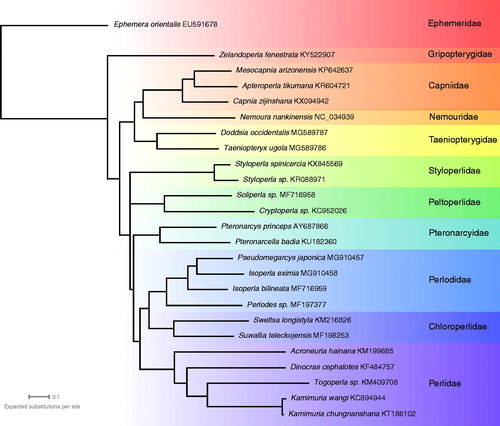
We obtained a mitochondrial genome of 16,385 bp, (average coverage 98×) Genbank Accession KY522907. The Z. fenestrata mitogenome retains the typical insect mitogenome gene set, including 13 protein-coding genes (ND1-6, COX1-3, ND4L, ATP8, ATP6, and CYTB), 22 tRNA genes, two ribosomal RNAs and a control region. The phylogenetic tree reinforces the believed deep divergence between Antarctoperlaria and Arctoperlaria ().
Discussion
This mitogenome confirms the placement of Antarctoperlaria as basal and divergent from Arctoperlaria (Chen and Du Citation2018) – which had previously been suggested both from morphology (Zwick Citation2000) and from COI sequences (McCulloch et al. Citation2016). As this is the first mitogenome from any member of Antarctoperlaria, this sequence should be of interest for phylogenetic reconstructions within Polyneoptera, and for looking at mitochondrial evolutionary rates within Plecoptera.
Acknowledgements
The authors wish to thank Aaron Jeffs, NZGL and Darren Hart for their contributions in helping to analyze and sequence the genome.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stdler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bethoux O, Cui YY, Kondratieff B, Stark B, Ren D. 2011. At last, a Pennsylvanian stem-stonefly (Plecoptera) discovered. Bmc Evol Biol. 11:248.
- Chen Z-T, Du Y-Z. 2018. The first two mitochondrial genomes from Taeniopterygidae (Insecta: Plecoptera): structural features and phylogenetic implications. Int J Biol Macromol. 111:70–76.
- Cui YY, Bethoux O, Kondratieff B, Shih CK, Ren D. 2016. The first fossil salmonfly (Insecta: Plecoptera: Pteronarcyidae), back to the Middle Jurassic. Bmc Evol Biol. 16:217.
- Dussex N, Chuah A, Waters JM. 2016. Genome-wide SNPs reveal fine-scale differentiation among wingless alpine stonefly populations and introgression between winged and wingless forms. Evolution. 70:38–47.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:13.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMB net J. 17:10–12.
- McCulloch GA, Wallis GP, Waters JM. 2009. Do insects lose flight before they lose their wings? Population genetic structure in subalpine stoneflies. Mol Ecol. 18:4073–4087.
- McCulloch GA, Wallis GP, Waters JM. 2016. A time-calibrated phylogeny of southern hemisphere stoneflies: testing for Gondwanan origins. Mol Phylogenet Evol. 96:150–160.
- McLellan ID. 1977. Alpine and southern Plecoptera from New Zealand, and a new classification of gripopterygidae. N Z J Zool. 4:119–147.
- McLellan ID. 1999. A revision of Zelandoperla Tillyard (Plecoptera: Gripopterygidae: Zelandoperlinae). N Z J Zool. 26:199–219.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25:126–127.
- Sinichenkova ND. 1997. Palaeontology of stoneflies. In: Landolt P, Sartoni M, Fribourg M, editors. Ephemeroptera and Plecoptera: biology–ecology–systematics. Fribourg, Switzerland: Mauron, Tinguely and Lachat SA; p. 561–565.
- Veale AJ, Foster BJ, Dearden PK, Waters JM. 2018. Genotyping-by-sequencing supports a genetic basis for alpine wing-reduction in a New Zealand stonefly. Scientific Rep. 8:16275.
- Zwick P. 2000. Phylogenetic system and zoogeography of the plecoptera. Annu Rev Entomol. 45:709–746.