Abstract
Among all species of flies, Xylophagidae is one of the most primitive Brachycera families. We sequenced and annotated the mitochondrial genome of Heterostomus sp., the first representative of subfamily Xylophaginae with complete mitochondrial data. This mitogenome is 15,897 bp totally, which consists of 22 transfer RNAs, 13 protein-coding genes, two ribosomal RNAs and non-coding control regions. All genes have the conservational arrangement like other published species of Brachycera. The nucleotide composition biases toward A and T, the overall A + T% was up to 78.7% of the entire mitogenome. Both Bayesian inference and ML analysis strongly supported the monophyly of Xylophagidae. Our results also suggested that Xylophagomorpha is the sister group to Stratiomyomorpha.
Xylophagidae, usually known as awl-flies, contains over 100 species which predominantly distributed in Nearctic and Palaearctic regions (Ovtshinnikova Citation2006). Adults usually occur in wooded and forested areas, particularly near water. The larvae are generally considered to be predacious, which live under bark and in decaying wood (McAlpine et al. Citation1981).
Xylophagidae is the sole member of the infraorder Xylophagomorpha, which presently contains three subfamilies (Woodley Citation2011). However, there is no general agreement about the systematic placement of Xylophagomorpha and relationships within Xylophagidae (Yeates Citation2002; Wiegmann et al. Citation2011; Lambkin et al. Citation2013; Shin et al. Citation2018). The system of three subfamilies, Xylophaginae, Coenomyiidae and Rachiceridae, had been controversial for a long time, especially as for Coenomyidae. (Coulson Citation1965; McAlpine et al. Citation1981).
The only one published mitochondrial genome representing Xylophagidae is from subfamily Coenomyiinae (Wang et al. Citation2016), so we provide another complete mitochondrial genome of Heterostomus sp. from subfamily Xylophaginae for further multiple phylogenetic analysis. Specimens were collected at Calaveras Big Trees State Park, CA, the DNA and specimens are deposited in the Entomological Museum of China Agricultural University. The complete data has been submitted to NCBI database with the accession number MH817480.
The sequencing has followed the procedures of Gillett et al. (Citation2014), the pooled dsDNA sample was sent to BIONONA CO., LTD for library construction and sequenced by the Illumina HiSeq2500 platform. The final filtered reads were assembled with Meta-IDBA (Peng et al. Citation2012).
The complete mitochondrial genome of Heterostomus sp. is 15,897 in length and consist of 37 canonical mitochondrial genes and one non-coding control region. The overall nucleotide composition of Heterostomus sp. mitochondrial genome was 39.2% of A, 39.5% of T, 12.2% of C, and 9.1% of G. The nucleotide composition of control region showed high bias toward A and T which the A + T% is up to 90.2%.
The ATG was the most popular start codon shared with ATP6, CO2, CO3, CYTB, ND2, ND4, ND4L, and start codon ATT was shared with ATP8, ND3, ND5, ND6. Particularly, the CO1 begins with codon CCG, and the ND1 begins with codon ATA. The conservative stop codon TAA was shared with most of the PCGs except for two genes, CO1 were terminated with an incomplete stop codon T, while the gene ND5 was ended with TA.
Figure 1. Phylogenetic tree of 10 Brachyceran species which consist of two Xylophagidae species. Bayesian posterior probabilities and ML bootstrap values were labeled at each node.
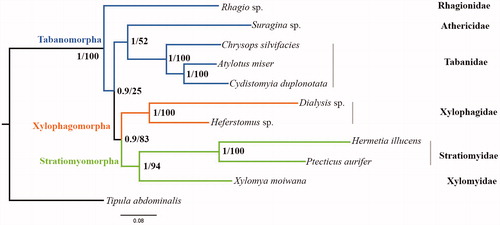
Phylogenetic trees were inferred using two approaches (BI on MrBayes v3.2.6 (Ronquist et al. Citation2012), and ML on RAxML-HPC2 v8.2.10 (Stamatakis Citation2006) for dataset containing 13 PCGs and two ribosomal RNA genes (). Tipula abdominalis was chosen as outgroup. The monophyly of the Xylophagomorpha and Stratiomyomorpha were consistently supported, and the sister relationship between Xylophagomorpha and Stratiomyomorpha was supported in relatively high value which is same with recent mitochondrial (Wang et al. Citation2016) and morphological (Lambkin et al. Citation2013) studies. The monophyletic Xylophagidae in these analyses provide new phylogenetic evidence based on mitochondrial genome data: despite the divergence of adult morphology and larval feeding habits, the subfamilies Xylophaginae and Coenomyiinae should be united into the family Xylophagidea (Palmer and Yeates Citation2000).
Acknowledgements
The authors express our sincere thanks to Dr Shaun L. Winterton for collecting and identifying the specimens.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Coulson JRS. Alan (1965). A catalog of the Diptera of America north of Mexico. In E. R. D. United States, editors, U.S.: U.S. Govt. Print; p.296–299.
- Gillett CPDT, Crampton-Platt A, Timmermans MJTN, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of Weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237.
- Lambkin CL, Sinclair BJ, Pape T, Courtney GW, Skevington JH, Meier R, Wiegmann BM. 2013. The phylogenetic relationships among infraorders and superfamilies of Diptera based on morphological evidence. System Entomol. 38:164–179.
- McAlpine JPBV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM. 1981. In: M. T. J., editor, Manual of Nearctic Diptera, Vol. 1. Quebec, Canada: Research Branch of Agriculture; p.489–493.
- Ovtshinnikova OG. 2006. Musculature of the male genitalia of Xylophagus cinctus (De Geer) and relationships within the superfamily Xylophagoidea (Diptera, Brachycera). Entmol Rev. 86:709–716.
- Palmer CM, Yeates DK. 2000. Phylogenetic importance of immature stages: solving the riddle of Exeretonevra Macquart (Diptera: Xylophagidae). Ann Entomol Soc Am. 93:15–27.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. System Biol. 61:539–542.
- Shin S, Bayless K, Winterton S, Dikow T, Lessard B, Yeates D, Wiegmann B, Trautwein M. 2018. Taxon sampling to address an ancient rapid radiation: a supermatrix phylogeny of early brachyceran flies (Diptera). System Entomol. 43:277–289.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang K, Li X, Ding S, Wang N, Mao M, Wang M, Yang D. 2016. The complete mitochondrial genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with mitochondrial genome phylogeny of lower Brachycera (Orthorrhapha). Gene. 586:184–196.
- Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, et al. 2011. Episodic radiations in the fly tree of life. Proc Natl Acad Sci USA. 108:5690–5695.
- Woodley NE. 2011. A world catalog of the Xylophagidae (Insecta: Diptera). In: Thompson FC, Brake I, Lonsdale O, editors, Contributions to the Biosystematic Database of World Diptera. Vol.12. Moscow: Myia; p.522–568.
- Yeates DK. 2002. Relationships of extant lower Brachycera (Diptera): a quantitative synthesis of morphological characters. Zoologica Scripta. 31:105–121.
