Abstract
Fraxinus pennsylvanica is distributed in the central and eastern USA and Canada. It is assessed as Critically Endangered (CR). In this study, the complete chloroplast (cp) genome sequence of F. pennsylvanica was determined using next-generation sequencing. The entire cp genome was determined to be 155,543 bp in length. It contained large single-copy (LSC) and small single-copy (SSC) regions of 86,389 and 17,806 bp, respectively, which were separated by a pair of 25,674 bp inverted repeat (IR) regions. The genome contained 132 genes, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall AT content of the genome is 37.8%. A phylogenetic tree reconstructed by 10 chloroplast genomes reveals that F. pennsylvanica is most related with F. chiisanensis.
The genus Fraxinus L. comprises 43 species occurring in temperate and subtropical regions of the northern hemisphere (Miller Citation1995, Wallander Citation2008). Fraxinus pennsylvanica is suffering the devastating impact of a invasive pest and assessed as Critically Endangered (CR) under criteria A3e + 4ae (The IUCN Red List and Threatened Specis, 2017). This species is distributed in the central and eastern USA and Canada. It consists of at least three different ecotypes (Wallander Citation2008). It is often confused with other Fraxinus species. So, it is necessary to develop genomic resources for F. pennsylvanica to provide intragenic information for clarifying the taxonomic identities and valuable information about the course of evolution of Fraxinus.
The total genomic DNA was extracted from the fresh leaves of F. pennsylvanica (Shanghai Binhai Forest Park, 30.96°N, 121.91°E) using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). The DNA was stored at –20 °C in our lab until use. The whole genome sequencing was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq 2500 Sequencing System (Illumina, Hayward, CA, US). The filtered sequences were assembled with reference sequence of Fraxinus chiisanensis (GenBank: NC_037171) using the program NOVOPlasty (Dierckxsens et al. Citation2017). Annotation was performed using the DOGMA (http://dogma.ccbb.utexas.edu/) and BLAST searches. The plastome of F. pennsylvanica was determined to comprise double stranded, circular DNA of 155,543 bp containing two inverted repeat (IR) regions of 25,674 bp each, separated by large single-copy (LSC) and small single-copy (SSC) regions of 86,389 and 17,806 bp, respectively (NCBI acc. no. MH836622). The genome contained 132 genes, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The seven protein-coding genes, seven tRNA genes and four rRNA genes were duplicated in IR region. Fifteen genes contained one intron and two genes (clpP and ycf3) contained two introns. The overall GC content of F. pennsylvanica cp genome is 37.8% and the corresponding values in LSC, SSC and IR regions are 35.8%, 32.0%, and 43.2%, respectively.
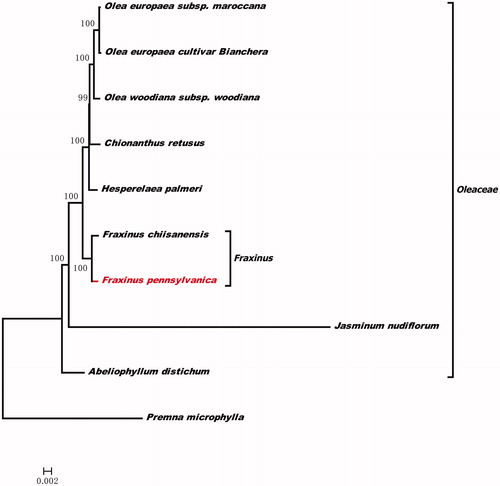
To investigate its taxonomic status, a maximum likelihood (ML) was reconstructed based on 9 complete chloroplast genomes sequences in Oleaceae (using the outgroup Premna microphylla) by FastTree version 2.1.10 (Price et al. Citation2010). The ML phylogenetic tree shows that F. pennsylvanica is most related with F. chiisanensis in Oleaceae, with bootstrap support values of 100% ().
Figure 1. The phylogenetic tree based on the nine complete chloroplast genome sequences. Accession numbers: Olea europaea cultivar Bianchera (NC013707), Olea europaea subsp. maroccana (NC015623), Olea woodiana subsp. woodiana (NC015608), Chionanthus retusus (NC035000), Hesperelaea palmeri (NC025787), Jasminum nudiflorum (NC008407), Abeliophyllum distichum (NC031445), Fraxinus chiisanensis (MG594385.1) and Premna microphylla (NC026291).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Miller GN. 1955. The genus Fraxinus, the ashes, in North America, North of Mexico. Cornell Univ Agric Exp Station Memoir. 335:1–64.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PloS ONE. 5:e9490.
- Wallander E. 2008. Systematics of Fraxinus (Oleaceae) and evolution of dioecy. Plant Syst Evol. 273:25–49.
