Abstract
Cercopithecus erythrotis camerunensis a subspecies of Cercopithecus erythrotis, endemic primate to West Africa, is listed as a vulnerable species under the IUCN red list, due to a rapid population decline resulting from intense hunting, bushmeat trade and habitat loss. In this study, the complete mitochondrial genome of C. erythrotis camerunensis was sequenced and characterized using next-generation sequencing technique. The total length of the complete mitogenome is 16,645 bp, 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes (tRNAs) and a non-coding control region (D-loop region). Its efficient protection from anthropogenic activities has been a major challenge, thus, the complete mitochondrial genome sequence reported here will give molecular information for future evolutionary research and scientific basis for its conservation.
Cercopithecus erythrotis camerunensis is a small, colourful monkey with distinctive facial markings, native to Nigeria-Cameroon border, West Africa (Oates et al. Citation2008: Kingdon et al. Citation2013). Unfortunately, C. erythrotis camerunensis is threatened by deforestation, hunting and bushmeat trade (Fa et al. Citation2000: Linder Citation2008; Oates et al. Citation2008) and has thus led to its population decline. This species has been classified as a Vulnerable species by the International Union for Conservation of Nature (IUCN) on the ‘Red List of Threatened Species’, which makes the protection and conservation of this important species pertinent.
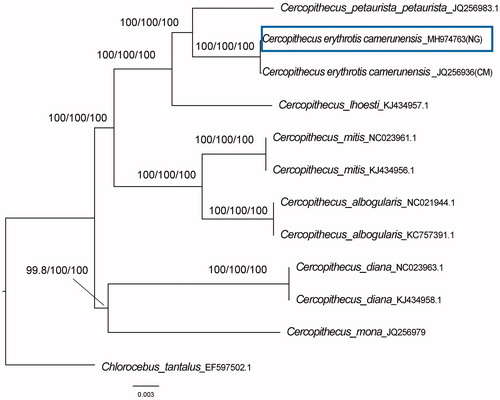
Figure 1 Phylogenetic relationships among Cercopithecus based on mtDNA genome of 11 sequences with their species name and accession numbers, Chlorocebus tantalus (EF597502.1) was set as outgroup taxon. Cercopithecus erythrotis camerunesnsis with accession number (JQ256936) was collected from Cameroon (CM) while the new sequence of Cercopithecus erythrotis camerunensis (MH974763) was collected from Nigeria (NG). Number above each node indicates the NJ, ML and MP bootstraps support values, respectively.
In this study, the complete mitochondrial genome of C. erythrotis camerunensis was sequenced. Animal collection permits and ethical approval were obtained from National Park Service, Abuja, Nigeria. This permit and licenses are necessary for collection of samples from the park and transportation of the samples from Nigeria to China. The tissue sample of C. erythrotis camerunensis was collected from carcass exhibit seized from poachers in Cross River National Park, Nigeria, West Africa. It was deposited in the Animal Branch of Germplasm Bank of Wild species, Kunming Institute of Zoology, Chinese Academy of Sciences. DNA was extracted using DNeasy Blood & Tissue Kit (QiaGen, Valencia, CA). Library construction and sequencing was done in the Southern China DNA Barcoding Center, with Illumina Miseq platform. Assembling was carried out with SPAdes (http://cab.spbu.ru/software/spades/). Reads were assembled using Linux-OS SPAdes genome assembler v3.12.0 (Bankevich et al. Citation2012) with k-mer 21, 33, 55. The tRNAs sequences were confirmed using online Search Service tRNAscan-SE (Schattner et al. Citation2005).
We obtained the complete mtDNA genome of an individual of C. erythrotis camerunensis (MH974763). The genome organization consists of 37 genes (13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and one control region (D-loop)). Most of the PCGs initiation codons were ATG except for ND3 and ND2 with slight differences in the initiation codon of ATT and ATA respectively. The mitogenome was 16,645 bp in length, with an overall base composition of A: 31.8%, C:29.7%, G:12.8% and T:25.8%. Nucleotide composition was estimated by MEGA 7.0 (Kumar et al. Citation2016). In addition, nine of the 13 protein-coding genes had complete termination codon TAG, AGG, TAA or GAA, while COIII, ND4, ND3 and CYTB genes terminate with incomplete stop codon (T-). A total of 22 gaps/overlaps were identified among the genes. It has the same gene arrangement and similar codon usage with other Cercopithecidae mitochondrial genome (Li et al. Citation2009; Lei et al. Citation2010; Chang et al. Citation2014; Wang et al. Citation2014).
To validate the reliability of the sequence, comparison was made with other mitochondrial sequences from Cercopithecus species. The phylogenetic position was estimated from complete mtDNA sequences (). The newly characterized sequence had 100% bootstrap (). The neighbor joining tree (NJ) and maximum likelihood (ML) were performed in MEGA 7.0 while maximum parsimony (MP) was estimated using PAUP* (V4.0) (Swofford Citation2003).
This study will give a useful database for analyzing the phylogenetic relationship of C. erythrotis camerunensis and other Cercopithecus species. This will provide valuable scientific insights on its conservation and restoration.
Acknowledgements
The authors are grateful to the Nigeria National Park Service Commission, Abuja, Nigeria for their support. This work was supported by Kunming Institute of Zoology, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chang P, Li Y, Zhang Z, Hwang D. 2014. Complete mitochondrial genome of sooty mangabey, Cercocebus atys atys (Mammalia: Primates: Cercopithecidae). Mitochondrial DNA Part A. 27(6):3897–3893. doi: 10.3109/19401736.2014.987253.
- Fa JE, Garcia Yuste JE, Castelo R. 2000. Bush meat markets on Boiko Island as a measure of hunting pressure. Conserv Biol. 14:1602–1613.
- Kingdon J, Happold D, Butynski T, et al. 2013. Mammals of Africa, Vols. 1–6. UK: A and C Black. pp. 373–375.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lei R, Shore GD, Brenneman RA, Engberg SE, Sitzmann BD, Bailey CA, Kimmel LM, Randriamampionona R, Ranaivoarisoa JF, Louis EE, et al. 2010. Complete sequence and gene organization of the mitochondrial genome for Hubbard's sportive lemur (Lepilemur hubbardorum). Gene. 464:44–49.
- Li M, Zhou Y, Feng G, Su SB. 2009. The critical role of Toll-like receptor signaling pathways in the induction and progression of autoimmune diseases. Curr Mol Med. 9:365–374.
- Linder JM. 2008. The Impact of Hunting on Primates in Korup National Park, Cameroon: Implications for primate Conservation. PhD thesis. New York: City University of New York.
- Oates JF, Gippoliti S, Groves CP. 2008. Cercopithecus erythrotis. The IUCN Red List of Threatened Species (accessed 2018 Sept 21). https://www.iucnredlist.org/species/4218/10651543
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sunderland, MA: Sinauer Associates.
- Wang W, Liu JY, Wang HF, Yang MY, Liu QY, Ding MX. 2014. The complete mitochondrial genome of white tuted-ear marmoset, Callithrix jacchus (Primates: Callitrichinae). Mitochondrial DNA. 1:1–2. doi:10.3109/19401736.2014.971287.
