Abstract
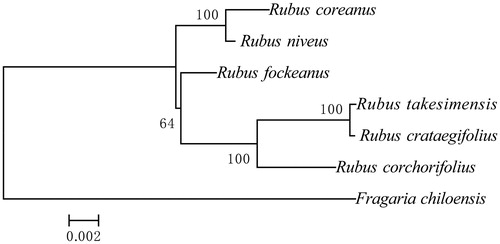
The wild Rubus species Rubus coreanus, which is widely distributed in east Asia, shows great promise as a genetic resource for breeding. The complete chloroplast genome of the species is 155,405 bp in length, with an 85,322 bp large single copy (LSC) and a 17,693 bp small single copy separated by two inverted repeats. The plastid encodes a total of 132 genes, including 87 protein-coding genes, 37 tRNA genes and eight rRNA genes. The average GC content of the protein coding genes is 39.7%. Maximum-likelihood (ML)-based phylogenetic analysis indicated that the R. coreanus has the closest relationship to R. nievus.
The genus Rubus L. has more than 1000 species widely distributed all over the world. The most common and economically important ones are those belong to the subgenera Rubus and Ideaobatus. The increasing threads posed by pathogens including various fungi and virus have brought enumerations loses in production (Daubeny and Pepin Citation1974; Pritts Citation2001). R. coreanus, a popular wide resource has been used as diseases-resistance donor for a long time in the Rubus breeding programs (Keep et al. Citation1977; Finn et al. Citation2002). Despite the release of the whole genome sequences of the first Rubus species R. occidentalis (VanBuren et al. Citation2016), we still lake genetic information for most of the other Rubus species. What makes it worse is the difficulty in species identification due to the ubiquitously existence of interspecies hybridization and apomixis (Amsellem et al. Citation2001).
Here in this study, the total genomic DNA of R. creanuse was extracted from fresh leaves collected in Ya'an, Sichuan Province, China (29°58'24.5''N, 103°00'18.7''E) using a modified protocol based on cetyltrimethyl ammonium bromide (CTAB) (Gawel and Jarret Citation1991). Voucher specimens of the species were deposited in the herbarium for horticultural plants, Sichuan Agricultural University (these herbaria were not indexed in Index Herebariorum) as described (Wang et al. Citation2016). Fifteen pairs of universal primers for the whole chloroplast genome developed by Zhang et al. (Citation2016) were used. Each segment of the genome was amplified in a Bio-rad PTC100 PCR system in a 200 μL tube containing 1.25U PrimeSTART GXL DNA polymerase (TaKaRa, Dalian, China), 1 × PrimSTAR GXL buffer, 1.6 mmol/L dNTP, 0.5 μmol/L each primer, and 100 ng genomic DNA. The PCR conditions were adjusted to give a sharp and unique target band. Thirty cycles were used for all amplifications. All PCR amplicons were purified using the E.Z.N.A DNA purification kits from OMEGA (Norcross, GA). Same quantities of each fragment were pooled and six micrograms of the DNA was used to construct 350 bp insertion libraries using the Illumina TruSeq DNA PCR-free library preparation protocol. The libraries were sequenced on a HiSeq X Ten platform (BGI, Shenzhen, China). All 150 bp Illumina paired-end (PE) reads were assembled by using Geneious Pro (v11.0) after quality control. The inverted repeats were judged based on the reads spanning and been manually edited if necessary. The complete chloroplast genome were annotated by using the GeSeq module (Tillich et al. Citation2017) of CHLOROBOX toolbox with default settings which integrated tRNAscan-SE, ARGAGORN and BLAT for tRNA and rRNA identification. The predicted annotation was verified using BLAST similarity search. All annotations were manually edited before submitting to GenBank. Finally, other six complete or partial chloroplast genome from Rubus were collected from public database. They are R. corchorifolius (KY419958.1), R. crataegifolius (MG189543.1), R. fockeanus (KY420018.1), R. niveus (KY419961.1) and R. takesimensis (NC_037991.1). Fragaria chiloenisis (NC_019601) was used as the outgroup. All genomes were aligned using progressive Mauve (Darling et al. Citation2004) to identify locally collinear blocks. The resulting fasta file was applied to infer the phylogeny in IQ-TREE v1.5.5 (Nguyen et al. Citation2015). Branch support was assessed using the fast bootstraping option implemented in the software.
The complete chloroplast genome (Accession no. MH992398) of the species is 155,405 bp in length, with an 85,322 bp large single copy (LSC) and a 17,693 bp small single copy separated by two inverted repeats (26,195 bp). The plastid encodes a total of 132 genes, including 87 protein-coding genes, 37 tRNA genes and eight rRNA genes. The average GC content of the genome is 37.3%, in comparing with that of the protein coding genes (39.7%). Maximum-likelihood (ML)-based phylogenetic analysis indicated that R. coreanus has the closest relationship with R. niveus (). The complete chloroplast genome will provide valuable molecular data for species identification and phylogenetic analysis for Rubus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Amsellem L, Noyer JL, Hossaert-McKey M. 2001. Evidence for a switch in the reproductive biology of Rubus alceifolius (Rosaceae) towards apomixis, between its native range and its area of introduction. Am J Bot. 88:2243–2251.
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403.
- Daubeny HA, Pepin HS. 1974. Variations among red raspberry cultivars and selections in susceptibility to the fruit rot causal organisms Botrytis cinerea and Rhizopus spp. Can J Plant Sci. 54:511–516.
- Finn C, Swartz H, Moore P, Ballington J, Kempler CJA. 2002. Use of 58 Rubus species in five North American breeding programmes-breeders notes. Acta Hortic. 585:113–119.
- Gawel NJ, Jarret RL. 1991. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep. 9:262–266.
- Keep E, Knight VH, Parker JH. 1977. Rubus coreanus as donor of resistance to cane diseases and mildew in red raspberry breeding. Euphytica. 26:505–510.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Pritts MP. 2001. From plant to plate: how can we redesign Rubus production systems to meet future expectations? Acta Hortic. 585:537–543.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- VanBuren R, Bryant D, Bushakra JM, Vining KJ, Edger PP, Rowley ER, Priest HD, Michael TP, Lyons E, Filichkin SA, et al. 2016. The genome of black raspberry (Rubus occidentalis). Plant J. 87:535–547.
- Wang Y, Chen Q, Chen T, Tang HR, Liu L, Wang XR. 2016. Phylogenetic insights into Chinese Rubus (Rosaceae) from multiple chloroplast and nuclear DNAs. Front Plant Sci. 7:968.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytem Evol. 54:219–227.