Abstract
Magnolia sinica, distributed in Yunnan province of China, is a critically endangered (CR) species based on the IUCN Red List. Here we provide the complete chloroplast (cp) genome sequence of M. sinica. Its length is 159,443 bp with four sub-regions: 88,156 bp of LSC region and 18,748 bp of SSC region are separated by two IR regions (each 26,570 bp). The genome contains 79 CDSs, four rRNAs, and 30 tRNAs. Phylogenetic analysis of cp genome of M. sinica with previously reported chloroplast genomes shows that M. yunnanensis is a sister to M. sinica with high bootstrap value.
As a species belonging to Magnoliaceae, Magnolia sinica (Y. W. Law) Noot. is one of flagship species for conservation in China (Wang et al. Citation2016). It is categorized as Critically Endangered (CR) species in the IUCN Red List based on its restricted distributions: only 52 individuals in eight localities of Yunnan province in China have been reported (Wang et al. Citation2016). And also, the species has been targeted as one of the 120 PSESP (Plant Species with Extremely Small Populations) priorities for urgently rescuing in China (Chen et al. Citation2016). Diagnostic characters of this taxon in Magnoliaceae include opened prefoliations and ventrally dehiscing carpels (Figlar and Nooteboom Citation2004). Taxonomic position of this taxon in Magnoliaceae has long been controversial. It has been considered as an independent genus, Manglietiastrum Y. W. Law, in Magnoliaceae. However, its taxonomic position has been changed to a section included in subgenus Talauma (Nooteboom Citation1985) in Magnolia, a section included in Manglietia (Liang and Nooteboom Citation1993), or a section included in subgenus Gynopodium in Magnolia (Figlar and Nooteboom Citation2004). A recent molecular phylogenetic study on Magnoliaceae using 10 chloroplast (cp) DNA regions has recognized 11 major clades in subgenus Magnolioideae (Kim et al. Citation2001; Kim and Suh Citation2013).
In this study, we report a complete sequence of cp genome of M. sinica (GenBank accession no. NC023241). We extracted DNA from a leaf sample of a tree cultivated in Kunming Institute of Botany, which is propagated from a seed of the wild tree (voucher: S. Kim 1098 deposited in the herbarium of the Natural Products Research Institute in the Seoul National University, Korea).
To complete the cp genome of M. sinica, we amplified and sequenced 134 PCR fragments covering the entire cp genome region using 268 primers reported previously (Song et al. Citation2018b). PCR conditions of our experiment followed those of Kim et al. (Citation2011). PCR products were purified with commercial PCR product purification kit (PCR SV; GeneAll, Seoul, Korea), and sequenced with an ABI 3700 sequencer (ABI, Carlsbad, CA) in both directions. We aligned all sequences from PCR products using Sequencher (ver. 4.9; Gene Code Co., Ann Arbor, MI). The completed cp genome was annotated using DOGMA (Wyman et al. Citation2004).
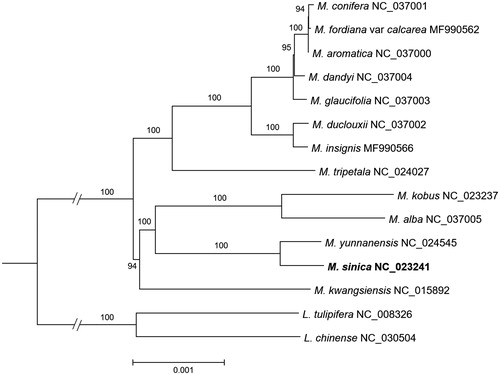
The cp genome of M. sinica is 160,044 bp, containing four subregions: 88,156 bp of large single copy region and 18,748 bp of small single copy region are separated by two inverted repeat regions (each 26,570 bp). The number of identified genes is 113, including 79 CDSs, four rRNAs, and 30 tRNAs. Inverted repeat regions contain 17 genes (six protein-coding genes, four rRNAs, and seven tRNAs). Phylogenetic analysis was performed for 14 previously reported cp genomes in Magnoliaceae and cp genome of M. sinica using raxmlGUI (version 1.3; Silvestro and Michalak Citation2012) with GTR + G + I model and 500 bootstrap replications (). Results of analysis demonstrate that M. sinica is a sister to M. yunnanensis with 100% of bootstrap value. This study will play an important role in conservation and restoration studies on M. sinica, a critically endangered species in Magnoliaceae.
Figure 1. A maximum-likelihood tree constructed for 14 cp genomes from previously reported cp genomes in Magnoliaceae (Cai et al. Citation2006; Kuang et al. Citation2011; Yang et al. Citation2014; Li et al. Citation2016; Zhu et al. Citation2016; Wang et al. Citation2017; Song et al. Citation2018a; Zheng and Xu Citation2018) and the cp genome of Magnolia sinica. The tree is rooted by two species of subfamily Liriodendroideae. Numbers above the node indicate bootstrap value (values over 50% are indicated).
Acknowledgments
This work was supported by the National Research Foundation of Korea [NRF-2017R1D1A1B03034952] to S. Kim; the National Science Foundation in China-Yunnan [U1302262] to W. Sun.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77.
- Chen Y, Chen G, Yang J, Sun WB. 2016. Reproductive biology of Magnolia sinica (Magnoliaecea), a threatened species with extremely small populations in Yunnan, China. Plant Diversity. 38:253–258.
- Figlar RB, Nooteboom HP. 2004. Notes on Magnoliaceae IV. Blum – J Plant Tax Plant Geog. 49:87–100.
- Kim S, Park CW, Kim YD, Suh Y. 2001. Phylogenetic relationships in family Magnoliaceae inferred from ndhF sequences. Am J Bot. 88:717–728.
- Kim S, Suh Y. 2013. Phylogeny of Magnoliaceae based on ten chloroplast DNA regions. J Plant Biol. 56:290–305.
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 54:663–673.
- Li B, Li Y, Cai Q, Lin F, Meng Q, Zheng Y. 2016. The complete chloroplast genome of a Tertiary relict species Liriodendron chinense (Magnoliaceae). Conservation Genet Resour. 8:279–281.
- Liang CB, Nooteboom HP. 1993. Notes on Magnoliaceae III: The Magnoliaceae of China. Ann Missouri Botanical Garden. 80:999–1104.
- Nooteboom H. 1985. Notes on Magnoliaceae. Blumea. 31:65–87.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Song E, Park S, Park J, Kim S. 2018a. The chloroplast genome sequence of Magnolia kobus DC. (Magnoliaceae). Mitochondrial DNA B Resour. 3:342–343.
- Song E, Park S, Sun W, Kim S. 2018b. A set of primers for studying chloroplast genomes in Magnoliaceae. Appl Plant Sci. Submitted.
- Wang B, Ma Y, Chen G, Li C, Dao Z, Sun W. 2016. Rescuing Magnolia sinica (Magnoliaceae), a critically endangered species endemic to Yunnan, China. Oryx. 50:446–449.
- Wang G, Hou N, Zhang S, Luo Y. 2017. Characterization of the complete chloroplast genomes of seven Manglietia and one Michelia species (Magnoliales: Magnoliaceae). Conserv Genet Resour. 9:1–4.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Zheng W, Xu X. 2018. The complete chloroplast genome of endangered Manglietia insignis, a rare landscaping tree with red lotus-like flowers. Conservation Genet Resour. 10:27–30.
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209:1747–1756.
