Abstract
We determined the complete mitochondrial (mt) sequence of the Cocktail wrasse, Pteragogus flagellifer. The mt genome is 16,807 bp long and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two noncoding regions. All its mt protein-coding genes begin with an ATG start codon except for CO1, which begins with GTG. The order of the mt genes is largely identical to that of typical vertebrates, although the position of the TMF-regulated nuclear protein (trnP) is unique among known Labridae gene arrangements. The phylogenetic tree of Labridae showed that P. flagellifer is relatively close to Pseudolabrus.
Pteragogus is a genus of wrasses native to the Indo-West Pacific, the Red Sea to South, Papua New Guinea, northern to southern Japan, the shores of the Korean Peninsula, and south to Australia. Cocktail wrasse, Pteragogus flagellifer, is found among coral or rocky reefs and weedy marine bottoms. Jeju Island is rich in coral and rocky reefs and, therefore, is the habitat of numerous wrasses including P. flagellifer.
Pteragogus flagellifer was caught in a coastal reef off Jeju Island, South Korea (geospatial coordinates: 33°31'59"N, 126°35'23"E). The total DNA was extracted and stored in deep freezer in Jeju Biodiversity Research Institute. The complete mitochondrial (mt) genome was sequenced (GenBank accession num. EF409976). The mt genome was 16,807 bp long, consisting of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and two noncoding regions.
The A/C/G/T base composition of the 13 PCGs was 22.3/30.8/18.0/28.9. The PCG proportion of G at the third position in a codon is relatively low, and pyrimidines are overrepresented in the second codon position (68.0%), as previously reported for other vertebrate mt genomes, owing to the hydrophobic character of the proteins (Naylor et al. Citation1995). Among 22 tRNAs, trnSAGY has no discernible dihydrouridine (DHU) stem, similar to that in the lamprey (Lee and Kocher Citation1995), bichir (Noack et al. Citation1996), ayu (Ishiguro et al. Citation2001), and rock bream (Oh et al. Citation2007).
The P. flagellifer mt gene arrangement is identical to that of typical vertebrates, although the trnP position relative to the control region is unique among known Labridae gene arrangements.
The major control region of the P. flagellifer mt genome spans 893 bp, consistent with those of other vertebrates (Lee et al. Citation1995). The control region is located between the trnT and trnP genes while other typical bony fishes have their control region located between the trnP and trnF genes. Many mt genome studies have revealed numerous independently evolved gene rearrangements (Kumazawa et al. Citation1996; Macey et al. Citation1997), and mt gene rearrangements related to the control region have been studied in various animals (Ishikawa et al. Citation2000; Sumida et al. Citation2001; Kumazawa and Endo Citation2004; Dong and Kumazawa Citation2005; Peng et al. Citation2006; Su et al. Citation2007; Jühling et al. Citation2012). This discovery might provide meaningful insights for studying the speciation and evolutionary process of Labridae.
The mt genome of P. flagellifer has an additional presumably redundant, non-coding region located between trnP and trnF. The non-coding region is 232 bp long and contains several palindromic sequences.
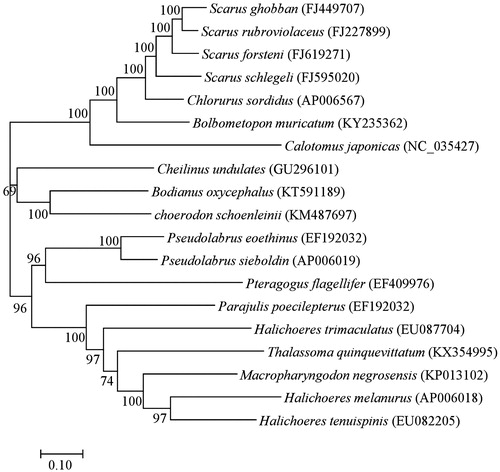
To examine P. flagellifer in the molecular phylogeny of Labridae, we constructed a maximum-likelihood (ML) tree using the nucleotide sequences of 13 PGGs from all Labridae mt genomes registered in GenBank (). In the ML tree, P. flagellifer was a sister taxon to Pseudolabrus. The tree topology was not consistent with that of previous studies (Cowman et al. Citation2009).
Figure 1. Molecular phylogenetic analysis of mitochondrial (mt) 13 protein-coding genes (PCGs) in 19 species of Labridae. The phylogenetic tree was constructed using maximum-likelihood (ML) method based on the GTR + G + I (Nei and KumarCitation2000) as the best nucleotide substitution model based on Akaike information criterion (AIC) (AkaikeCitation1974). The rate variation model allowed for some sites to be evolutionarily invariable. Percentages of trees where associated taxa were clustered together were shown next to branches. Evolutionary analyses were conducted using the MEGA7 (Kumar et al.Citation2016).
Disclosure statement
The authors declare that they have no competing interests.
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Autom Control. 19:716–723.
- Cowman PF, Bellwood DR, van Herwerden L. 2009. Dating the evolutionary origins of wrasse lineages (Labridae) and the rise of trophic novelty on coral reefs. Mol Phylogenet Evol Phylogenet Evol. 52:621–631.
- Dong S, Kumazawa Y. 2005. Complete Mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 61:12–22.
- Ishiguro N, Miya M, Nishida M. 2001. Complete mitochondrial DNA sequence of ayu Pleoglossus altivelis. Fisheries Sci. 67:474–481.
- Ishikawa S, Kimura Y, Tokai T, Tsukamoto K, Nishida M. 2000. Gene rearrangement around the control region in the mitochondrial genome of conger eel Conger myriaster. Fisheries Sci. 66:1186–1188.
- Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. 2012. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40:2833–2845.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kumazawa Y, Endo H. 2004. Mitochondrial genome of the komodo dragon: efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 11:115–125.
- Kumazawa Y, Ota H, Nishida M, Ozawa T. 1996. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 13:1242–1254.
- Lee WJ, Kocher TD. 1995. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome: early establishment of the vertebrate genome organization. Genetics. 139:873–887.
- Lee WJ, Conroy J, Howell WH, Kocher TD. 1995. Structure and evolution of teleost mitochondrial control regions. J Mol Evol. 41:54–66.
- Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. 1997. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol. 14:91–104.
- Naylor GJ, Collins TM, Brown WM. 1995. Hydrophobicity and phylogeny. Nature. 373:555–556.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York, NY: Oxford University Press.
- Noack K, Zardoya R, Meyer A. 1996. The complete mitochondrial DNA sequence of the bichir (Polypterus ornatipinnis), a basal ray-finned fish: ancient establishment of the consensus vertebrate gene order. Genetics. 144:1165–1180.
- Oh DJ, Kim JY, Lee JA, Yoon WJ, Park SY, Jung YH. 2007. Complete mitochondrial genome of the rock bream Oplegnathus fasciatus (Perciformes, Oplegnathidae) with phylogenetic considerations. Gene. 392:174–180.
- Peng QL, Nie LW, Pu YG. 2006. Complete mitochondrial genome of Chinese big-headed turtle, Platysternon megacephalum, with a novel gene organization in vertebrate mtDNA. Gene. 380:14–20.
- Su X, Wu XB, Yan P, Cao SY, Hu YL. 2007. Rearrangement of a mitochondrial tRNA gene of the concave-eared torrent frog, Amolops tormotus. Gene. 394:25–34.
- Sumida M, Kanamori Y, Kaneda H, Kato Y, Nishioka M, Hasegawa M, Yonekawa H. 2001. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of the Japanese pond frog Rana nigromaculata. Genes Genet Syst. 76:311–325.
