Abstract
Metapenaeus is one of the most important genera of Penaeidae. However, the systematics and phylogenetics studies of family Penaeidae have so far been limited. In this study, we report the complete mitochondrial genome sequence of M. affinis. The mitogenome has 16,027 base pairs (65.9% A + T content) and made up of a total of 37 genes (13 protein-coding, 22 transfer RNAs and two ribosomal RNAs), and a putative control region. This study was the second available complete mitogenomes of Metapenaeus and will provide useful genetic information for future phylogenetic and taxonomic classification of Penaeidae.
Decapoda marine shrimp, especially Penaeid shrimp is valuable resources for fisheries and aquaculture in both tropical and subtropical regions(Calo-Mata et al. Citation2009). Jinga shrimp (Metapenaeus affinis) is an Indo-West Pacific species of family Penaeidae, which inhabits in the depth 5–90 m and prefers mud or sandy-mud bottoms. The identification and characterization of Penaeid shrimp traditionally relied on morphometric analysis. However, the phenotypic similarities of shrimps and the environmental influence of morphometric characteristics make taxonomic identification difficult (Fernandes et al. Citation2017). Moreover, there are still many controversial issues on systematics and phylogenetics of Penaeidae in recent years (Zhang et al. Citation2016). The complete mitochondrial genome is using molecular techniques for better understanding of genome evolution and phylogeny. Here, we report the second complete mitochondrial genome sequence of genus Metapenaeus, which will provide a better insight into systematics and phylogenetics of Penaeid.
Muscle samples of M. affinis from three individuals were collected from Guangxi province, China (Qinzhou, 21.604014 N, 108.685301 E), and the whole body specimen (#GQ0203) were deposited at Marine biological Herbarium, Guangxi Institute of Oceanology, Beihai, China. The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq NanoTM kit (Illumina, San Diego, CA) and were sequenced (2 × 150 bp paired-end) using HiSeq platform at Novogene Company, China. Mitogenome assembly was performed by MITObim (Hahn et al. Citation2013). The complete mitogenome of M. ensis (GenBank accession number: NC_026834) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS (Bernt et al. Citation2013).
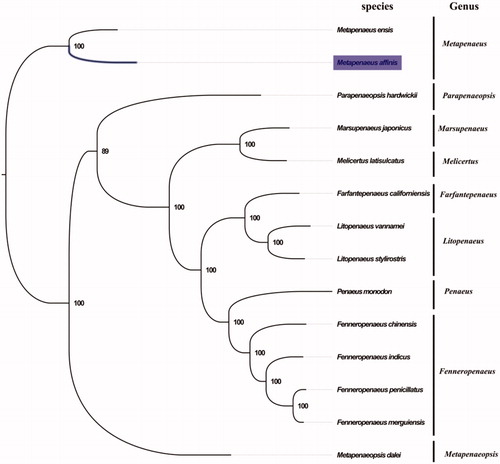
The complete mitogenome of M. affinis was 16,027 bp in length (GenBank accession number: MG815825), and containing the typical set of 13 protein-coding, 22 tRNA and two rRNA genes, and a putative control region. The overall base composition of the mitogenome was estimated to be A 34.6%, T 31.3%, C 21.5% and G 12.6%, with a high A + T content of 65.9%, which is similar, but slightly higher than Parapenaeopsis hungerfordi (64.5%) (Zhong et al. Citation2018). The gene arrangement in M. affinis is identical to that found in M. ensis, which is in the same order as the pancrustacean ground pattern (Zhang et al. Citation2016). Furthermore, the result of phylogenetic tree of 14 species (including other 13 species from family Penaeidae in NCBI) also supported the monophyletic status of Metapenaeus (), more mitogenome of Metapenaeus are desirable to further understand the phylogenetics of Penaeidae. The complete mitochondrial genome sequence of M. affinis was the second sequenced mitogenomes within the genus Metapenaeus, which will contribute to further phylogenetic and taxonomic studies for the family Penaeidae.
Figure 1. Phylogenetic tree of 14 species in family Penaeidae. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 100 bootstrap replicates. The bootstrap values were labeled at each branch nodes. The gene's accession number for tree construction is listed as follows: Metapenaeus ensis (NC_026834), Parapenaeopsis hardwickii (NC_030277), Marsupenaeus japonicus (NC_007010), Melicertus latisulcatus (MG821353), Farfantepenaeus californiensis (NC_012738), Litopenaeus vannamei (NC_009626), Litopenaeus stylirostris (NC_012060), Penaeus monodon (NC_002184), Fenneropenaeus chinensis (NC_009679), Fenneropenaeus indicus (NC_031366), Fenneropenaeus penicillatus (NC_026885), Fenneropenaeus merguiensis (NC_026884), and Metapenaeopsis dalei (NC_029457).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Calo-Mata P, Pascoal A, Fernández-No I, Böhme K, Gallardo JM, Barros-Velázquez J. 2009. Evaluation of a novel 16S rRNA/tRNAVal mitochondrial marker for the identification and phylogenetic analysis of shrimp species belonging to the superfamily Penaeoidea. Anal Biochem. 391:127–134.
- Fernandes TJR, Silva CR, Costa J, Oliveira MBPP, Mafra I. 2017. High resolution melting analysis of a COI mini-barcode as a new approach for Penaeidae shrimp species discrimination. Food Control. 82:8–17.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Zhang DC, Liu TT, Gong FH, Jiang SG. 2016. Shotgun assembly of the first mitochondrial genome of Metapenaeus (Metapenaeus ensis) with phylogenetic consideration. Mitochondrial DNA Part A. 27:3689–3690.
- Zhong SP, Zhao, YF, Zhang, Q. 2018. The complete mitochondrial genome of Parapenaeopsis hungerfordi (Decapoda: Penaeidae). Mitochondrial DNA Part B. 3:1212–1213.
