Abstract
Sunan yak is one of the unique and indigenous yak species in China. The complete mitochondrial genome sequence of the Sunan yak was first determined in this study. The mitochondrial DNA is a circular molecule with 16,322 bp in length, containing 12 protein-coding genes, two rRNA genes, 22 tRNA genes and a noncoding control region (D-loop), showed a typical mammal pattern. The overall nucleotide composition is A (33.71%), T(27.26%), C(25.82%), G(13.21%) with an A + T content of 60.97%. Phylogenetic analysis based on the mitochondrial genomes of 12 related species using MEGA 5.0 indicated that Sunan yak was most closely related to Qinghai Plateau yak and Maiwa yak.
Sunan yak is mainly spread in Sunan Yugu Autonomous County of Gansu Province, characterized by a strong adaptation to extreme high-altitude environment and good meat quality (Liu Citation2011). The complete mitochondrial genome sequence and structure has been widely applied as a powerful molecular tool to phylogenetic and population genetic studies and biodiversity conservation (Stephen Citation2014). However, the complete mitochondrial DNA sequences of Sunan yak have not yet been reported. In this study, we determined the complete mitochondrial DNA sequence of Sunan yak. The complete mitochondrial of Sunan yak would provide a detailed genetic information for the genetic structure study of this species.
Specimens of Sunan yak were obtained from Sunan Yugu Autonomous County, Gansu Province, China (E 99°36′51ʺ, N38°50′11ʺ). Muscle tissues were fixed in 95% ethanol and preserved in Key Laboratory of Yak Breeding Engineering of Gansu Province. Total genomic DNA was extracted from the skeletal muscle tissue using the animal tissue DNA isolation kit (ZDGSY, Beijing, China) according to the manufacturer’s instructions. Six pairs of primers were used to amplify the PCR products for sequencing and the sequencing results were assembled by using DNAStar 5.01 software. The complete mitogenome data of the Sunan yak was deposited in GenBank with an accession number: MH921427. The locations of protein-coding genes were determined by comparing with the corresponding sequences of other yak species.
The complete mitochondrial genome of Sunan yak is 16,322 bp in length, and consisted of 37 mitochondrial genes in total, including 13 protein-coding genes (COX1-3, CYTB, NAD1-6, NAD4L, ATP6 and ATP8), 22 tRNA genes, two ribosomal RNA subunits genes (12S and 16S rRNA) and a major non-coding control region (D-Loop region). The arrangement of these genes is conserved as other yaks in mammalian (Wang et al. Citation2011; Guo et al. Citation2016). There are three overlaps between protein-coding genes in the same strand: ATP8 overlaps with ATP6 for 40 bp, ND4L overlaps with ND4 for 7 bp, and ND5 overlaps with ND6 for 7 bp. The overlap of the ATPase genes appears to be common in most vertebrate mitochondrial genome (Broughton et al. Citation2001; Li et al. Citation2018). From the base composition analysis, the percent A + T content is 60.97% (33.71% for A, 27.26% for T, 25.82% for C and 13.21% for G).
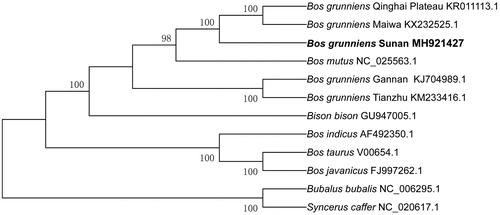
To validate the phylogenetic position of Sunan yak, a phylogenetic tree based on 12 mitochondrial protein-coding genes of Bovidae was constructed by MEGA 5.0 software (Tamura et al. Citation2011) with neighbor-joining (NJ) tree using 1000 bootstrap replicates. According to phylogenetic tree analysis, 12 species are clustered into two orders, and Sunan yak has a closer genetic relationship with Qinghai Plateau yak and Maiwa yak (). In this study, we provided the mitogenome sequence of Sunan yak, which may help in taxonomic classification, phylogenetic reconstruction and conservation strategies of the endemic species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Guo X, Pei J, Bao P, Chu M, Wu X, Ding X, Yan P. 2016. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondrial DNA A. 27:2889–2890.
- Li Y, Cao K, Fu C. 2018. Ten fish mitogenomes of the tribe Gobionini (Cypriniformes: Cyprinidae: Gobioninae). Mitochondrial DNA B. 3:802–803.
- Liu J. 2011. Research on Yak meat Maturation Mechanism and Meat Quality after Slaughter. [MA.Sc. Thesis]. Lanzhou (Gansu): Gansu Agricultural University.
- Stephen L. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang Z, Yonezawa T, Liu B, Ma T, Shen X, Su J, Guo S, Hasegawa M, Liu J. 2011. Domestication relaxed selective constraints on the yak mitochondrial genome. Mol Biol Evol. 28:1553–1556.