Abstract
Petunia hybrida is a popular ornamental plant and a model for chloroplast research. A complete P. hybrida chloroplast genome sequence would advance phylogenetic studies in the Solanaceae sub-family Petunioideae and is essential for developing synthetic biology tools to study and manipulate chloroplast function for fundamental and industrial applications. Here we present the 156,575 bp P. hybrida chloroplast genome sequence. It is comprised of 87,083 bp large single copy and 18,630 bp small single copy regions separated by 25,431 bp inverted repeats. Seventy-nine proteins, 30 transfer-RNAs and four ribosomal RNA species are encoded by 113 unique genes. The results of phylogenetic analysis using complete Solanaceous chloroplast genomes are consistent with other data that supports an early split for the P. hybrida lineage away from the branch giving rise to groups containing major crops such as tobacco (Nicotianoideae) and potato (Solanoideae).
Petunia hybrida is the world’s most popular bedding plant (Van der Krol and Immink Citation2016) and a model for genetic research (Gerats and Vandenbussche Citation2005). P. hybrida belongs to the Solanaceae, a family with major crops including tobacco (Nicotiana tabacum), tomato (Solanum lycopersicon) and potato (Solanum tuberosum). P. hybrida has a rich history of chloroplast research (Bovenberg et al. Citation1981; Palmer and Thompson Citation1982; Cornu and Dulieu Citation1988). Recently, P. hybrida chloroplast transformants (Zubko et al. Citation2004) provided the first experimental verification of enhanced containment of chloroplast transgenes in the field (Horn et al. Citation2017). The P. hybrida sequence reported here represents the first complete chloroplast genome (cp-genome) sequence for a member of the Petunioideae. This facilitates phylogenetic studies of this sub-family (Ando et al. Citation2005) as well as advancing chloroplast transformation technologies in P. hybrida (Avila and Day Citation2014).
P. hybrida var. Pink Wave was obtained from Thompson and Morgan, Ipswich, UK (52°02′44.5″N, 1°05′50.0″E) and stored in the Plant Science Laboratories, Manchester University (53°27′51.9″N 2°13′46.0″W). DNA extracted from Percoll-gradient-purified chloroplasts (Primavesi et al. Citation2008) was sequenced using 300 bp paired end reads (Nextera DNA kit libraries) on the Illumina MiSeq platform (San Diego, USA). De novo assembly of the 156,575 bp cp genome (GenBank Accession MF459662) used a rbcL seed sequence (Aldrich et al. Citation1986) and NOVOPlasty v 2.5.9 (Dierckxsens et al. Citation2017). Average coverage was over 10,000 reads (Geneious R11, Biomatters Ltd, Auckland). Annotation used GeSeq (Tillich et al. Citation2017), CpGAVAS (Liu et al. Citation2012) followed by manual curation with Vector NTI advance v9 (Invitrogen, Carlsbad).
The 156,575 bp P. hybrida cp genome has the typical inverted repeat (IR) organisation of land plants (Palmer and Thompson Citation1982) and coding content of Solanaceae cp-genomes (Amiryousefi et al. Citation2018). Two 25,431 bp IRs separate an 87,083 bp large single copy region from a 18,630 bp small single copy region. It contains 113 unique genes encoding 79 proteins, 30 transfer-RNAs and 4 ribosomal RNA (rRNA) genes. The rps12 gene is trans-spliced and the psbL and ndhD genes contain ACG start codons consistent with RNA editing.
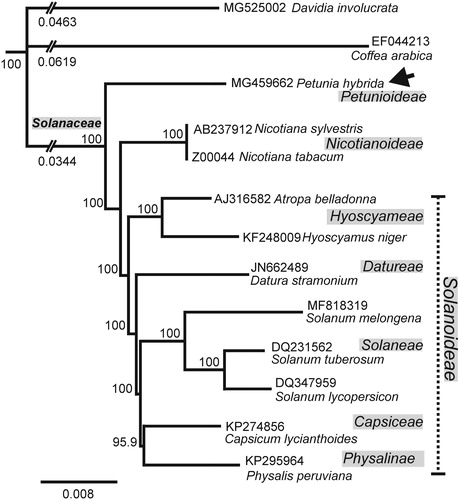
Complete cp-genomes were aligned using MAFFT (Katoh and Standley Citation2013) in Geneious R11 after removing one copy of the large inverted repeat. Geneious R11 tree builder used neighbour joining (Tamura and Nei Citation1993) and Davidia involucrata as the outgroup. The resulting tree () shows the Petunioideae branching away from the group that contains the Nicotianodeae and Solanoideae. This is consistent with the Petunia lineage splitting from the branch giving rise to the Nicotianoideae and Solanoideae about 30 million years ago (Bombarely et al. Citation2016). High conservation of P. hybrida and N. tabacum cp-genomes (98% base identity) is consistent with successful transplantation of P. hybrida chloroplasts into N. tabacum (Glimelius and Bonnett Citation1986) and the use of N. tabacum chloroplast sequences to target integration of transgenes in P. hybrida chloroplasts (Zubko et al. Citation2004). The P. hybrida cp-genome sequence provides a template for editing the P. hybrida cp-genome by homologous recombination (Avila et al. Citation2016).
Figure 1. Neighbour joining consensus tree of indicated chloroplast genomes. Bootstrap values (1000 replicates) are indicated at nodes. Distances shown for truncated branches. The position of P. hybrida MF459662 (this work) is arrowed. Sequences retrieved from GenBank. Scale bar: substitutions per site.
Acknowledgements
We thank the Genomics Technologies Next Generation Sequence Facility and Richard Leong in research IT for their support, and Genbank for sequences used in the Phylogenetic Tree.
Disclosure statement
The authors alone are responsible for the content and writing of the article. None of the authors report any conflict of interest.
Additional information
Funding
References
- Aldrich J, Cherney B, Merlin E, Palmer J. 1986. Sequence of the rbcL gene for the large subunit of ribulose bisphosphate carboxylase-oxygenase from Petunia. Nucleic Acids Res. 14:9534–9534.
- Amiryousefi A, Hyvonen J, Poczai P. 2018. The chloroplast genome sequence of bittersweet (Solanum dulcamara): plastid genome structure evolution in Solanaceae. Plos One. 13:e96069. doi:10.1371/journal.pone.0196069
- Ando T, Kokubun H, Watanabe H, Tanaka N, Yukawa T, Hashimoto G, Marchesi E, Suarez E, Basualdo IL. 2005. Phylogenetic analysis of Petunia sensu Jussieu (Solanaceae) using chloroplast DNA RFLP. Ann Bot. 96:289–297.
- Avila EM, Day A. (2014) Stable plastid transformation of petunia. In: Maliga P, editor. Chloroplast biotechnology: methods and protocols (Methods in Molecular Biology) vol 1132. New York (NY): Humana Press; p. 277–293.
- Avila EM, Gisby MF, Day A. 2016. Seamless editing of the chloroplast genome in plants. BMC Plant Biol. 16:6.
- Bombarely A, Moser M, Amrad A, Bao M, Bapaume L, Barry CS, Bliek M, Boersma MR, Borghi L, Bruggmann R, et al. 2016. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat Plants. 2(6):74.
- Bovenberg WA, Kool AJ, Nijkamp HJJ. 1981. Isolation, characterization and restriction endonuclease mapping of the Petunia hybrida chloroplast DNA. Nucleic Acids Res. 9:503–517.
- Cornu A, Dulieu H. 1988. Pollen transmission of plastid-DNA under genotypic control in Petunia hybrida hort. J Hered. 79:40–44.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:955. doi:10.1093/nar/gkw955
- Gerats T, Vandenbussche M. 2005. A model system comparative for research: Petunia. Trends Plant Sci. 10:251–256.
- Glimelius K, Bonnett HT. 1986. Nicotiana cybrids with Petunia chloroplasts. Theor Appl Genet Appl Genet. 72:794–798.
- Horn P, Nausch H, Baars S, Schmidtke J, Schmidt K, Schneider A, Leister D, Broer I. 2017. Paternal inheritance of plastid-encoded transgenes in Petunia hybrida in the greenhouse and under field conditions. Biotechnol Rep. 16:26–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 29:537–550.
- Primavesi LF, Wu HX, Mudd EA, Day A, Jones HD. 2008. Visualisation of plastids in endosperm, pollen and roots of transgenic wheat expressing modified GFP fused to transit peptides from wheat SSU RubisCO, rice FtsZ and maize ferredoxin III proteins. Transgenic Res. 17:529–543.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Van der Krol AR, Immink RGH. 2016. Secrets of the world's most popular bedding plant unlocked. Nat Plants. 2:82.
- Zubko MK, Zubko EI, van Zuilen K, Meyer P, Day A. 2004. Stable transformation of petunia plastids. Transgenic Res. 13:523–530.
