Abstract
In the study, the complete mitogenome sequence of the Chilin fish (Varicorhinus macrolepis) has been sequenced. The mitochondrial DNA is 16,594 nucleotides and contains two ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, and one control region, along with gene order and organization, being similar to most of the other related fish mitogenome of the databases. The phylogenetic analysis showed V. macrolepis is close to genus Poropuntius and Spinibarbus. The results can provide a basic database for the evolutionary analysis for the V. macrolepis.
The Chilin fish (Varicorhinus macrolepis) is an ancient species of Cyprinidae, which only distributed in the rivers of Mountain Tai. The established research revealed that Chilin fish did not belong to genus Varicorhinus, which against the former morphological data (Qu et al. Citation2017). The fish has been recognised as a National Second-class Protected Animal by the List of Aquatic Wild Animal Protection in China (Chen Citation2007).
Here, we presented the complete mitochondrial genome of V. macrolepis (GenBank Accession Number MH924834). The specimen was obtained from the river in Shangliyuan Village, Tai’an, Shan Dong province, China. (36.15°N, 117.09°E) and stored in Specimens Laboratory, in Taishan Medical University under the accession number SL2016017. The DNA was extracted from muscle by using TIANamp Genomic DNA Kit following the manufacturer’s instructions (Tiangen Inc., Beijing, China). 15 pairs of primers were designed based on the conserved sequence of relative species. The complete mitogenome has 21 transfer RNA genes, 13 protein-coding genes, 2 ribosomal RNA genes and one control region (D-Loop).
The total base composition of the mitogenome is A (31.23%), T (24.54%), C (28.02%) and G (16.21%). The percentage of AT 55.77% was higher than GC (44.23%) rate, which shows AT bias. The character was generally in accordance with other fish mitochondrial genomes that in previous study (Chai et al. Citation2016).
All other genes are encoded on the heavy strand except for eight tRNA genes (tRNA-Gln, tRNAPro, tRNA-Glu, tRNA-Cys, tRNA-Ala, tRNA-Asn, tRNA-Tyr and tRNA-Ser) and the ND6 gene. All the protein-coding genes share the start codon ATG, except for CO1 gene (GTG start codon) and ND5 gene (CTG start codon). The stop codon, TAA, is present in ND1, CO1, ATP8, ND4L, ND5 and ND6 genes.
The 12S and 16S ribosomal RNA genes are 956 and 1675 bp. The genes were separately encoded between tRNA-Phe and tRNA-Val, tRNA-Val and tRNA-Leu. The length of D-loop is 939 bp, ranging from 15,656 to 15,654 bp.
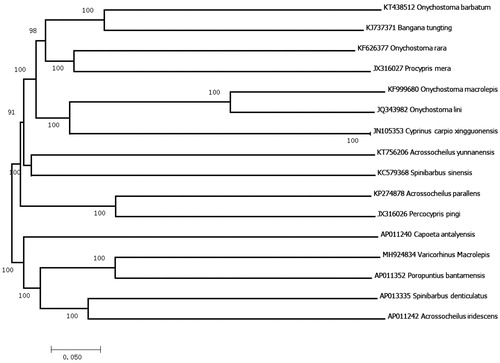
To validate the phylogenetic position of V. macrolepis, we used MEGA7 software (Kumar et al. Citation2016) to construct a neighbour-joining (NJ) with 1000 bootstrap replicates (Saitou and Nei Citation1987) containing complete mitogenomes of 16 species (). The figure showed that V. macrolepis clustered with P. bantamensi and S.denticulatus, which is consistent with previous report (Qu et al. Citation2017). The newly determined mitogenome provides important molecular data for phylogenetic analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chai A, Jie Z, Cui Q, Yuan C. 2016. Mitochondrial genome of onychostoma macrolepis (osteichthyes: cyprinidae). Mitochondrial DNA. 27:240–241.
- Chen C. 2007. The ecological habits and development, and utilization of Onychostoma macrolepis. Shandong Fish. 24:39.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Qu XJ, Li ZY, Song Q, Wang MW, Zhao BS. 2017. Mitochondrial partial DNA sequence analysis of Chi-lin fish Varicorhinus macrolepis from Taishan Mountain: implication for its taxonomy and evolution. J Shandong Univ Technol. 27:18–23.