Abstract
Plantago lagopus L. is the economically important medicine species plant of the family Plantaginaceae. Plantago lagopus L. is annual or perennial herbs, which is used as whole plants and seeds. Plantago lagopus L. has been used the effect of lowering blood fat, regulating blood sugar and diuresis. In this study, the complete chloroplast genome of Plantago lagopus L. was determined by the Illumina Hiseq4000 sequencing. The complete chloroplast genome of Plantago lagopus L. was 145,936 bp in length and displays a typical quadripartite structure of the large (LSC, 86,835 bp) and small (SSC, 18,431 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 20,335 bp each). It harbours 127 functional genes, including 84 protein-coding genes, 35 transfer RNA and 8 ribosomal RNA genes species. The overall nucleotide composition was: 30.4% A, 31.3% T, 19.4% C and 18.9% G, with a total G + C content of 38.3%. Phylogenetic relationship analysis shows that Plantago lagopus L. closely related to Plantago maritima L.
Plantago lagopus L. is a common herbal medicine belonging to the Plantaginaceae family and is native to China, India, other East Asia countries and South America (Rahn 1996). Seeds and husks of Plantago lagopus L. has been used extensively for the effect of lowering blood fat, regulating blood sugar and diuresis. Leaves of Plantago lagopus L. also consumed as a kind of food or tea. (Galisteo et al. Citation2005). At present, there are few studies on the genome and transcriptome of P. ovata. In this study, we used the Illumina Hiseq4000 sequencing to get the complete chloroplast genome of Plantago lagopus L., in order to provide information for the study of the origin Medicinal plants of P. lagopus L. in evolution, and also important to ensure the accuracy of clinical medication and the development and utilization of resources.
The specimen of Plantago lagopus L. was isolated from Jilin Agricultural University Jilin campus in Changchun, Jilin, China (125.41E; 43.81N) and the DNA of P. lagopus L. was stored in Jilin Agricultural University College of Life Science (No. JLAUCLS7). The P. lagopus L. DNA sample was sequenced as the paired end using the Hiseq4000 (Illumina Co., San Diego, CA). The P. lagopus L. complete chloroplast genome was preliminarily annotated using the Dual Organellar GenoMe Annotator (DOGMA) online program (Wyman et al. Citation2004), with default settings to identify protein-coding genes, rRNAs and tRNAs based on the Plant Plastid Code and BLAST homology searches. The secondary structures of transfer RNA (tRNA) genes were identified by using the ARAGORN (Laslett and Canback Citation2004) or through manually visual inspection.
The chloroplast genome sequence of P. lagoous L. is a closed-circular molecule of 145,936 bp in length, consisting of 86,835 bp LSC region and 18,431 bp SSC region separated by a pair of 20,335 bp IR region. It displayed 127 functional genes set which observed in this plant chloroplast, including 84 PCGs, 35 tRNA genes (one for each amino acid, two each for Alanine, Asparagine, Glycine, Methionine and Threonine, three each for Valine, Isoleucine, Arginine and Serine, four each for Leucine), and 8 genes for ribosomal RNA subunits (two each for rrn16, rrn23, rrn4.5 and rrn5). The annotated chloroplast genome of P. lagoous L. was submitted to GenBank of NCBI database under accession No.MH205736.
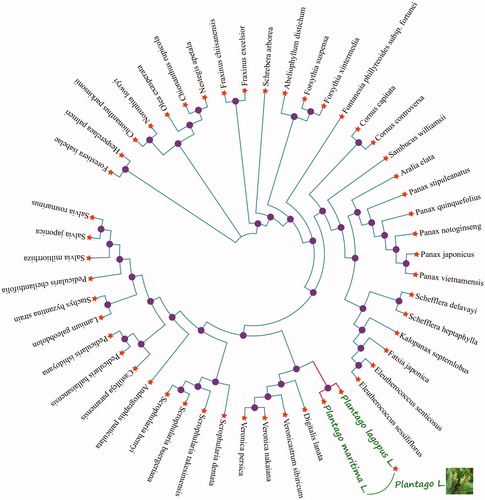
We selected other 48 related complete chloroplast genomes from GenBank to assess the phylogenetic relationship with P. lagopus L. between them. The genome-wide alignment of all plant complete chloroplast genomes was done by HomBlocks (Bi et al. Citation2017). The phylogenetic trees were reconstructed using maximum likelihood (ML) methods. ML phylogenetic tree was constructed using RaxML-8.2.4 (Stamatakis Citation2014) of which the bootstrap values were calculated using 10,000 replicates to assess node support. All the nodes were inferred with strong support by the ML methods. As shown in the phylogenetic tree (), Plantago lagopus L. showed the closest with Plantago maritima L.
Figure 1 Phylogenetic tree of Plantago lagopus L. and other 48 plant complete chloroplast genomes which yielded by Maximum-likelihood analysis. The phylogenetic tree was drawn without setting outgroup. All nodes exhibit above 90% bootstraps. The length of branch represents the divergence distance. The NCBI database accession number of Plantago lagopus L. to Plantago maritima L. in the counter clockwise direction is KT153019.1, JN637765.1, KR021045.1, KC456167.1, KT748629.1, KC456166.1, KU059178.1, KP036469.1, KP036468.1, KT028714.1, KY379906.1, KT153023.1, KX510276.1, MG525004.1, MG524998.1, MG255754.1, MG255756.1, MF579702.1, KT274029.1, MG255767.1, MG214254.1, MG594385.1, MG255758.1, MG255753.1, MG255766.1, MG255759.1, MG255752.1, LN515489.1, MG255755.1, KR232566.1, MG255762.1, KY646163.1, HF586694.1, KY751712.1, KY562590.1, KU170194.1, MG770330.1, KT959111.1, KF150644.2, MF861203.1, KP718626.1, KM590983.1, MF861202.1, KT724052.1, KT633216.1, KT724053.1, KY085895.1, KR297244.1.
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2017. HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Galisteo M, Sánchez M, Vera R, González M, Anguera A, Duarte J, Zarzuelo A, 2005. A diet supplemented with husks of Plantago ovata reduces the development of endothelial dysfunction, hypertension, and obesity by affecting adiponectin and tnf-alpha in obese zucker rats. J Nutr. 135:2399–2404.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Rahn KA. 1996. A phylogenetic study of the Plantaginaceae. Bot J Linn Soc. 120:145–198.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
