Abstract
The European fire-bellied toad, Bombina bombina, is a small aquatic toad belonging to the family Bombinatoridae. The species is native to the lowlands of Central and Eastern Europe, where population numbers have been in decline in recent past decades. Here, we present the first complete mitochondrial genome of the endangered European fire-bellied toad from Northern Germany recovered using iterative mapping. Phylogenetic analyses including other representatives of the Bombinatoridae placed our German specimen as sister to a Polish B. bombina sequence with high support. This finding is congruent with the postulated Pleistocene history of the species. Our complete mitochondrial genome represents an important resource for further population analysis of the European fire-bellied toad, especially those found within Germany.
The European fire-bellied toad (Bombina bombina) is a small- bodied toad and one of eight morphologically similar species in the genus Bombina. It is widely distributed throughout Eastern and Central Europe (Pabijan et al. Citation2013; Kuzmin et al. Citation2008). Despite this widespread distribution, isolated populations at the edge of its distribution range, as in Germany, have been in decline in recent decades due to intensive agriculture and ongoing habitat fragmentation (Arntzen Citation1978; Dolgener et al. Citation2012). This led the European fire-bellied toad to be listed as threatened with extinction on the National Red List in Germany (Beutler et al. Citation1998). Previous studies using complete mitochondrial genomes shed light on the phylogeny of the genus (Pabijan et al. Citation2013), but did not present individuals from the entire species range, including samples from only three central and southeast European localities (Poland, Turkey, and Austria). Northern German B. bombina populations are considered to be genetically depauperated and exhibit local Control Region haplotypes (Schröder et al. Citation2012), yet, the complete mitochondrial genome of the German B. bombina is still not available.
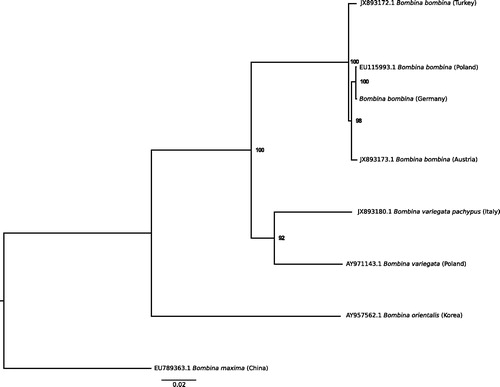
Our sample was a tadpole collected south of Neutestorf in Schleswig-Holstein, Germany (54°23′ N, 10°75′ E) and is permanently stored at the University of Potsdam. DNA and RNA were extracted using a customized Trizol/Chloroform protocol, built into an Illumina sequencing library using a NEXTflex Rapid Directional RNA-Seq Library Prep kit and sequenced on an Illumina HiSeq (Novogene, Hongkong). We trimmed adapter sequences using Cutadapt v.1.4 (Martin Citation2011) and removed PCR duplicates using Prinseq (Schmieder and Edwards Citation2011). We used iterative mapping to reconstruct the mitochondrial genome using MITObim v1.9 (Hahn et al. Citation2013) with default parameters and a mismatch value of 3. Mitochondrial genomes from B. bombina from three localities were used as bait sequences for three independent runs, Turkey (Genbank accession JX893172.1), Austria (JX893173.1) and Poland (EU115993.1). We constructed consensus sequences for each independent run using a 65% base call threshold and a minimum read depth of 10× in Geneious v8.1.9 (Kearse et al. Citation2012). We aligned the resultant consensus sequences using Mafft v7.271 (Katoh and Standley Citation2013) and called a final consensus sequence using a 75% base call threshold. We automatically annotated the resultant 17,381 bp genome (MH893761) using MITOS (Bernt et al. Citation2013) and found all protein-coding genes, tRNAs and rRNAs typical for vertebrate mitochondrial genomes. We aligned our sequence with a number of complete mitochondrial genomes from other Bombina species from Genbank using ClustalW (Thompson et al. Citation2002). We then performed a Maximum-Likelihood phylogenetic analysis in RaxML v.8 (Stamatakis Citation2014) with 500 bootstrap replicates, excluding the control region, and specifying B.maxima as outgroup. Results placed our sequence as sister to the Polish specimen with high confidence (bootstrap 100) (), a result consistent with the postglacial colonization history, as Poland and Germany is inhabited by the Northern lineage, Austria by the Southern lineage of colonization, and Turkey is close to the assumed glacial refugium (Hofman et al. Citation2007). This sequence represents an important resource for future studies on German Bombina bombina populations.
Acknowledgements
We are grateful to Arne Drews at the Landesamt für Landwirtschaft, Umwelt und ländliche Räume for his support regarding sampling permits and would also like to thank Moritz Ott for his assistance in the field.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arntzen JW. 1978. Some hypotheses on postglacial migrations of the fire-bellied Toad, Bombina bombina (Linnaeus) and the yellow-bellied Toad, Bombina variegata (Linnaeus). J Biogeogr. 5:339–345.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol Phylogenet Evol. 69:313–319.
- Beutler A, Geiger A, Kornacker PM, Kühnel KD, Laufer H, Podloucky R, Boye P, Dietrich E. 1998. Rote Liste der Kriechtiere (Reptilia) und Rote Liste der Lurche (Amphibia.). Rote Liste Gefährdeter Tiere Deutschlands Schriftenreihe Für Landschaftspflege Und Naturschutz. 55:48–52.
- Dolgener N, Schröder C, Schneeweiss N, Tiedemann R. 2012. Genetic population structure of the Fire-Bellied toad Bombina Bombina in an area of high population density: implications for conservation. Hydrobiologia. 689:111–120.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hofman S, Spolsky C, Uzzell T, Cogălniceanu D, Babik W, Szymura JM. 2007. Phylogeography of the Fire-Bellied toads Bombina: independent Pleistocene histories inferred from mitochondrial genomes. Mol Ecol. 16:2301–2316.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kuzmin SL, Pupina A, Pupins M, Trakimas G. 2008. Northern border of the distribution of the red-bellied toad (Bombina Bombina). Zeitschrift Für Feldherpetologie. 15:215–228.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12.
- Pabijan M, Wandycz A, Hofman S, Węcek K, Piwczyński M, Szymura JM. 2013. Complete mitochondrial genomes resolve phylogenetic relationships within Bombina (Anura: Bombinatoridae). Mol Phylogenet Evol. 69:63–74.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27:863–864.
- Schröder C, Pokorny I, Dolgener N, Herden C, Drews H, Tiedemann R. 2012. Allochthonous individuals in managed populations of the fire-bellied toad Bombina Bombina: genetic detection and conservation implications. Limnologica. 42:291–298.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics.