Abstract
In this study, the mitogenome of nine Raja pulchra individuals from Yellow Sea (n = 7) and even from Alaska (n = 2) was sequenced, and the genomic variation and phylogeny were examined. The length of the mitogenome was 16,906 and 16,907 bp showing 99.6% sequence similarity with the previously reported sequence. Of the one hundred and fifty-one single nucleotide polymorphisms, sixty-three were newly detected compared to previous studies. They were present in the 16s rRNA gene, 2 tRNA genes, 13 protein-coding genes, and a non-coding region. The mitogenomes and SNPs will provide a better understanding of the ecological genetics of R. pulchra.
Raja pulchra (mottled skate) is a species of Rajiformes, Rajidae. It is one of the largest fishes distributed in the Yellow Sea and the East Sea in the Korean Peninsula, and the Sea of Okhotsk from Japan to the coast of China (Ishihara Citation1990). Recently, it has been reported that R. pulchra in the Russian waters is found only in the coasts of Western Sakhalin (Antonenko et al. Citation2011). There has been a suggestion that R. pulchra should belong to the genus Beringraja, as they share the unique strategy of laying eggs (Ishihara et al. Citation2012; Chiquillo et al. Citation2014). The fish has been favoured by Koreans for a long time, and it is one of the foods cooked during important events. In the present study, we sequenced the mitochondrial DNA of nine individuals and examined variations in their mitogenome sequence from the reference sequence ((Jeong et al. Citation2016), Genbank Accession No. NC_025498.1).
Five fermented (HSF01-HSF05) and two non-fermented (DCNF01 and DCNF02) skates were obtained from Heuksan-do and Daechung-do, in the Yellow Sea, Republic of Korea, respectively. We also imported two fermented (USF01 and USF02) samples from Alaska, USA. Totally nine tissue samples were used for sequencing. Individual mitogenomes were sequenced according to the method described in our previous study (Kim and Lee Citation2016). Final assembled sequences were further annotated using a web-based automatic annotation server, MitoFish (Iwasaki et al. Citation2013). The complete sequence with annotations has been deposited in GenBank under the accession numbers MH844507-11 and MH899128-31. A phylogenetic tree was constructed with the Cox1 gene of 10 R. pulchra samples, including previously reported sequences, and 10 species belonging to the taxa Rajiformes. These sequences were aligned using ClustalW and used to construct the phylogenetic tree. Based on the reference sequence alignment, we were able to detect single nucleotide polymorphisms (SNPs). The polymorphic sites and amino acid sequences of protein-coding genes (PCGs) were collected to construct a phylogenetic tree using MEGA7 with 1000 bootstrap replications (Kumar et al. Citation2016).
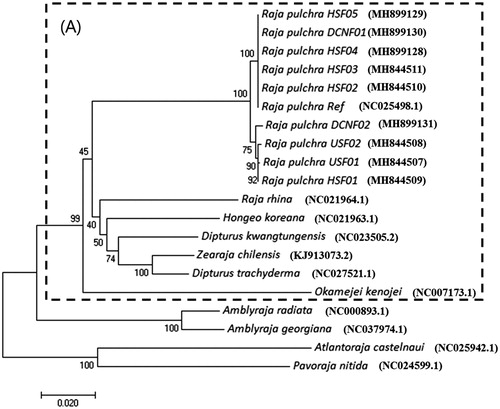
The mitogenome of nine individuals comprised the same contents and order of genes. The mitochondrial genome length and nucleotide composition were similar to those of the reference sequence. The length of the mitogenome was the same as that of the reference mitogenome (16,907 bp), but one nucleotide was deleted in four individuals (16,906 bp; USF01, USF02, HSF01, and DCNF02). The same site was deleted in these four individuals: the 2042nd nucleotide of the 16s rRNA gene. More SNPs were detected (A total of 151 SNPs, 0.893% of mitogenome sequence) than in previous study (Hwang et al. Citation2016). Among these newly detected 63 SNPs were located in 16s rRNA, 2 tRNA (tRNAGln and tRNAGlu), and 13 PCGs (Cox1-3, ND1-6, ND4L, ATP8, ATP6, and Cyt b). 12 SNPs in the six PCGs (ND1, ND2, ATP8, ND4, ND6, and Cyt b) were considered to be non-synonymous SNPs. The Cox1 gene is a good marker for classifying certain species. The phylogenic analysis using the Cox1 gene confirmed the current taxa of Rajiformes, Rajidae (). The skates inhabiting the Pacific Coast were clustered in the same group (). In the phylogenetic tree constructed using the SNPs and amino acids from the mitogenome sequence of 10 individuals (including the reference), the skates were not clustered by origin (Heuksan-do, Daecheung-do, and Alaska), but scattered. Raja rhina is a skate that lives around the Bering Sea and is distributed throughout from Alaska to California. Therefore, it was expected that USF01 and USF02 would be R. rhina, but they were found to be R. pulchra, like the Korean skate. The mitogenome analysis of the skates from Yellow Sea and Alaska provides a basic understanding of comparable information and ecological genetics of R. pulchra.
Disclosure statement
The authors declare that there are no conflicts of interest.
Additional information
Funding
References
- Antonenko DV, Solomatov SF, Balanov A, Tok KS, Kalchugin P. 2011. Occurrence of Skate Raja pulchra (Rajidae, Rajiformes) in Russian waters of the sea of Japan. J Ichthyol. 51:426.
- Chiquillo KL, Ebert DA, Slager CJ, Crow KD. 2014. The secret of the mermaid’s purse: phylogenetic affinities within the Rajidae and the evolution of a novel reproductive strategy in skates. Mol Phylogenet Evol. 75:245–251.
- Hwang JY, Jin G-D, Park J, Kim H, Lee C-K, Kwak W, Nam B-H, An CM, Park JY, Park K-H, et al. 2016. Complete genome sequence and SNPs of Raja pulchra (Rajiformes, Rajidae) mitochondria. Mitochondrial DNA A. 27:2975–2977.
- Ishihara H. 1990. The skates and rays of the western North Pacific: an overview of their fisheries, utilization, and classification. NOAA Technical Report NMFS. 90:485–498.
- Ishihara H, Treloar M, Bor PH, Senou H, Jeong C. 2012. The comparative morphology of skate egg capsules (Chondrichthyes: Elasmobranchii: Rajiformes). Bull Kanagawa Prefect Mus (Nat Sci). 41:9–25.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Jeong D, Kim S, Kim C, Myoung J, Lee Y. 2016. Complete mitochondrial genome of the mottled skate: Raja pulchra (Rajiformes, Rajidae). Mitochondrial DNA A. 27:1823–1824.
- Kim EB, Lee S. 2016. The complete mitochondrial genome of the Mongolian gerbil, Meriones unguiculatus (Rodentia: Muridae: Gerbillinae). Mitochondrial DNA A. 27:1457–1458.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.