Abstract
The complete mitochondrial genome of a cryptic species of tadpole shrimp from South Africa (Accession Number: MG770893) was recovered by low coverage shotgun sequencing. The mitogenome consists of 15,216 bp with a GC content of 30.95%. COX1 sequence alignment indicates that this specimen is the most closely related to Triops granarius with a pairwise nucleotide identity of less than 90% (86.6%). Phylogenetic analysis using whole mitogenome dataset supports its affiliation to the genus Triops with moderate support for its relatively basal position within the clade.
Tadpole shrimps belong to the genus Triops Schrank, 1803 (Schranck Citation1803) and are considered as ‘living fossils’ due to their conserved morphology and fossil records dating back to 250 MYA (Gore and Traverse Citation1986; Garrouste et al. Citation2009). Attempts to elucidate the phylogenetic relationships among members of the genus Triops, to date, have met with mixed success. Discordance between phylogenetic species and morphological species seems common (Suno-Uchi et al. Citation1997) and global genetic analysis of tadpole shrimps indicates high levels of cryptic diversity and unexpected relationships between major lineages (Vanschoenwinkel et al. Citation2012). Phylogeny inference using mitogenome sequences is emerging as a powerful tool to resolve shallow evolutionary relationships (Jacobsen et al. Citation2012; Gan et al. Citation2018). Despite the advent of next-generation sequencing platforms and reported successes of crustacean mitogenome assembly using established pipeline (Gan et al. Citation2014; Tan et al. Citation2015; Gan et al. Citation2016a), Triops mitogenomes remain scarce and lack representation from the African continent (Umetsu et al. Citation2002; Ryu and Hwang Citation2010; Horn and Cowley Citation2014; Gan et al. Citation2016b). We report the complete mitogenome of a cryptic species of tadpole shrimp collected from South Africa and assess its evolutionary relationships within the Triops genus.
A whole specimen of Triops sp. was collected from an ephemeral pond located near Grahamstown, South Africa (–33.423817; 26.448509), and stored in a 1.5 mL tube containing SDS-lysis solution and proteinase-K. A voucher specimen has been lodged at the Kansas Biological Survey (DCR-1128). The DNA extraction was performed from the lysate as previously described (Sokolov Citation2000). The gDNA was sheared to 500 bp using an ultrasonicator (Covaris, Woburn, MA), processed with NEBUltra DNA library preparation kit (New England Biolabs, Ipswich, MA) and sequenced on a MiSeq sequencer (Illumina, San Diego, CA). Annotation of mitogenome used MITObim v1.8 and MITOS v1, respectively (Bernt et al. Citation2013; Hahn et al. Citation2013). Thirteen protein-coding genes were extracted from each mitogenome, partitioned by codon position and aligned using TranslatorX (Abascal et al. Citation2010). The 12S and 16S ribosomal rRNA genes were extracted and aligned using Muscle v3.8.31 (Edgar Citation2004). The resulting 41 partitions (13 protein-coding genes ×3 codon position +2 rRNA genes) were trimmed with TriamAl v1.2 (‘-automated1 option’) (Capella-Gutiérrez et al. Citation2009). Model selection, partition merging, and maximum likelihood tree construction used IQTree v1.6.7 (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017).
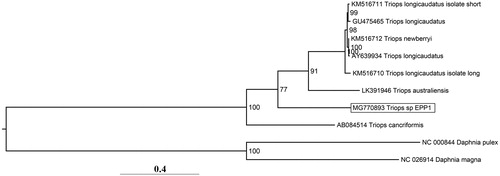
The mitogenome of Triops sp. EPP1 is 15,126 bp (30.95% GC content) with gene order that conforms to the primitive arrangement for Branchiopod crustaceans. A phylogenetic tree placed Triops sp. EPP1 within the Triops clade with moderate support for its basal position to all other Triops spp except Triops cancriformis (Bosc Citation1801) presumably due to an insufficient phylogenetic signal from uneven taxon sampling (). COX1 sequence analysis indicates that Triops sp. EPP1 is the most closely related to a T. granarius specimen from India (Accession code: HF911376, BOLD database as of 16 October 2018) with a nucleotide similarity of 86.6%, suggesting that it may be a cryptic species of T. granarius (Korn and Hundsdoerfer Citation2006).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38: W7–W13.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bosc LAG. 1801. Histoire naturelle des crustaces. Paris.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Gan HM, Schultz MB, Austin CM. 2014. Integrated shotgun sequencing and bioinformatics pipeline allows ultra-fast mitogenome recovery and confirms substantial gene rearrangements in Australian freshwater crayfishes. BMC Evol Biol. 14:19.
- Gan HM, Tan MH, Eprilurahman R, Austin CM. 2016a. The complete mitogenome of Cherax monticola (Crustacea: Decapoda: Parastacidae), a large highland crayfish from New Guinea. Mitochondrial DNA Part A. 27:337–338.
- Gan HM, Tan MH, Lee YP, Austin CM. 2016b. The complete mitogenome of the Australian tadpole shrimp Triops australiensis (Spencer & Hall, 1895)(Crustacea: Branchiopoda: Notostraca). Mitochondrial DNA Part A. 27:2028–2029.
- Gan HM, Tan MH, Lee YP, Schultz MB, Horwitz P, Burnham Q, Austin CM. 2018. More evolution underground: accelerated mitochondrial substitution rate in Australian burrowing freshwater crayfishes (Decapoda: Parastacidae). Mol Phylogenet Evol. 118:88–98.
- Garrouste R, Nel A, Gand G. 2009. New fossil arthropods (Notostraca and Insecta: Syntonopterida) in the continental Middle Permian of Provence (Bas-Argens Basin, France). Comptes Rendus Palevol. 8:49–57.
- Gore PJ, Traverse A. 1986. Triassic notostracans in the Newark supergroup, Culpeper Basin, northern Virginia. J Paleontol. 60:1086–1096.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Horn RL, Cowley DE. 2014. Evolutionary relationships within Triops (Branchiopoda: Notostraca) using complete mitochondrial genomes. J Crustac Biol. 34:795–800.
- Jacobsen MW, Hansen MM, Orlando L, Bekkevold D, Bernatchez L, Willerslev E, Gilbert MTP. 2012. Mitogenome sequencing reveals shallow evolutionary histories and recent divergence time between morphologically and ecologically distinct European whitefish (Coregonus spp.). Mol Ecol. 21:2727–2742.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587.
- Korn M, Hundsdoerfer AK. 2006. Evidence for cryptic species in the tadpole shrimp Triops granarius (Lucas, 1864) (Crustacea: Notostraca). Zootaxa. 1257:57–68.
- Lucas H. 1864. Bulletin entomologique. Séances de la Société entomologique de France. Séance du 24 Février 1864. Communications. Annales de la Société Entomologique de France, Series 4. 4:XI–XII.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ryu J-S, Hwang UW. 2010. Complete mitochondrial genome of the longtail tadpole shrimp Triops longicaudatus (Crustacea, Branchiopoda, Notostraca). Mitochondrial DNA. 21:170–172.
- Schranck FVP. 1803. Fauna boica. Landshut. 3:25.
- Sokolov EP. 2000. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J Molluscan Stud. 66:573–575.
- Suno-Uchi N, Sasaki F, Chiba S, Kawata M. 1997. Morphological stasis and phylogenetic relationships in Tadpole shrimps, Triops (Crustacea: Notostraca). Biol J Linn Soc. 61:439–457.
- Tan MH, Gan HM, Schultz MB, Austin CM. 2015. MitoPhAST, a new automated mitogenomic phylogeny tool in the post-genomic era with a case study of 89 decapod mitogenomes including eight new freshwater crayfish mitogenomes. Mol Phylogenet Evol. 85:180–188.
- Umetsu K, Iwabuchi N, Yuasa I, Saitou N, Clark PF, Boxshall G, Osawa M, Igarashi K. 2002. Complete mitochondrial DNA sequence of a tadpole shrimp (Triops cancriformis) and analysis of museum samples. Electrophoresis. 23:4080–4084.
- Vanschoenwinkel B, Pinceel T, Vanhove MPM, Denis C, Jocque M, Timms BV, Brendonck L. 2012. Toward a global phylogeny of the living fossil; crustacean order of the Notostraca. Plos One. 7:e34998.